Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.62 n.4 San José Oct./Dec. 2014
Epiphytic and endophytic bacteria that promote growth of ethnomedicinal plants in the subtropical forests of Meghalaya, India
Bacterias epífitas y endófitas que promueven el crecimiento de plantas etnomedicinales en los bosques subtropicales de Meghalaya, India
Bacterias epífitas y endófitas que promueven el crecimiento de plantas etnomedicinales en los bosques subtropicales de Meghalaya, India
Abstract
The present study was aimed to investigate the endophytic and epiphytic bacteria associated with selected ethnomedicinal plants from the pristine subtropical forests of Meghalaya and analyse them for plant growth promotion and antagonistic ability. This study is an attempt to explore plant associated bacteria which are beneficial to host plants, and thus aid in the conservation of ethnomedicinal plants of the studied subtropical forests, which are dwindling due to exploitation. The plant growth promotion parameters like indole acetic acid (IAA) production, mineral phosphate solubilisation, acid phosphatase activity, presence of 1-aminocyclopropane-1-carboxylic acid deaminase (ACC) gene, nitrogen fixation, cellulose digestion, chitin and pectin degradation were screened among the isolates. The study revealed significant differences in bacterial population not only between the epiphytic and endophytic microhabitats, but also amongst the host plants. Out of the 70 isolated plant associated bacteria, Bacillus sp., Serratia sp., Pseudomonas sp., Pantoea sp., and Lysinibacillus sp. showed potent plant growth promotion properties. Bacillus siamensis C53 and B. subtilis cenB showed significant antagonistic activity against the tested pathogens. This study indicated the isolates inhabiting the plants prevalent in the subtropical sacred forests could be explored for use as plant growth promoters while practising the cultivation and conservation of ethnomedicinal plants. Rev. Biol. Trop. 62 (4): 1295-1308. Epub 2014 December 01.
Key words: ethnomedicinal plants, endophytic and epiphytic bacteria, plant growth promotion, antagonistic.
Resumen
El presente estudio tuvo como objetivo investigar las bacterias endófitas y epífitas asociadas con plantas medicinales seleccionadas de los bosques subtropicales prístinos de Meghalaya, para estimular el crecimiento de plantas y la capacidad antagónica. Este estudio es un intento de explorar las plantas asociadas a bacterias que benefician a la planta hospedera, y así ayudar en la conservación de plantas medicinales de los bosques subtropicales estudiados, los cuales son cada vez más escasos debido a la explotación. Los parámetros de promoción de crecimiento de las plantas, tales como: índice de producción de ácido acético (IAA), solubilización de fosfato mineral, actividad de la fosfatasa ácida, presencia del gen 1-aminociclopropano-1-ácido carboxílico desaminasa (ACC), fijación de nitrógeno, digestión de celulosa, quitina y pectina fueron seleccionados entre los aislamientos. El estudio reveló diferencias significativas en la población bacteriana, no sólo entre los microhábitats epífitos y endófitos, sino también entre las plantas hospederas. De las 70 bacterias aisladas de plantas asociadas, Bacillus sp., Serratia sp., Pseudomonas sp., Pantoea sp. y Lysinibacillus sp. mostraron potentes propiedades estimulantes del crecimiento vegetal. Bacillus C53 siamensis y B. subtilis cenB mostraron actividad antagónica significativa contra los patógenos probados. Este estudio indicó que los aislamientos que habitan en las plantas predominantes en los bosques sagrados subtropicales podrían explorarse para su uso como promotores del crecimiento vegetal, mientras se practica el cultivo y conservación de plantas medicinales.
Palabras clave: Plantas meidicinales, bacterias endófitas y epífitas, de promoción del crecimiento de las plantas, antagonismo.
Ethnomedicinal plants are harvested and used for traditional medicinal practices by ethnic tribes living in and around the tropical and subtropical forests. Plants including herbs, shrubs and trees have been reported to be used as drugs or medicaments by the tribal people of North-Eastern region of India (Hynniewta & Kumar, 2008; Syiem, Sharma & Saio, 2009). An estimate of 80% of the population in developing countries rely on traditional medicines, mostly plant drugs, for their primary health care needs (Farnsworth, Akerele, Bingel, Soejarta, & Eno, 1985). There is an increasing demand for medicinal plants in both developed and developing countries due to growing recognition of natural products being non-toxic, having no side effects and availability at affordable prices (Sekar & Kandavel, 2010). Unfortunately, deforestation coupled with exploitation, has resulted in dwindling population of important medicinal plants. Plants maintain a complex ecosystem where microbial communities interact continu¬ously (Stierle, Strobel, & Stierle, 1993). Endophytes are specific group of microorganisms (bacterial and fungal) that can be found residing in internal healthy plant tissues without causing visible damage to their hosts (Petrini, 1991; Hallmann, Quadt-Hallmann, Mahaffee, & Kloepper, 1997), whereas, epiphytes colonizes plant surfaces and are in contact with the outer environment.
The study of plant-associated bacteria and their potential to confer plant beneficial effects are important not only for understanding their ecological role and their interaction with plants, but also for biotechnological applications. Recent studies have shown that plant growth promotion may be attributed to the secretion of plant growth promoting secondary metabolites by the associated microbes (Kobayashi & Columbo, 2000; Hamayun et al. 2010). Thus, investigating on plant-microbe interaction and assessing the role of indigenous pre-dominant culturable endophytic and epiphytic bacteria, associated with ethnomedicinal plants, for plant growth promotion and antagonistic activity, is required to maintain the production of medicinal plants for their novel natural products.
Materials and methods
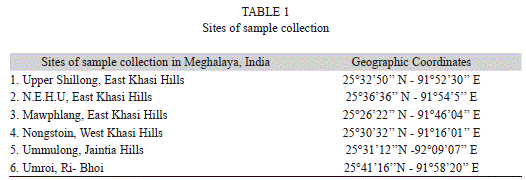
Plant sample collection: Healthy plants used by the traditional herbal practitioners were collected from different forests of Meghalaya, India (25°30’50” N - 90°12’11” E and 25°27’00” N - 92°12’00” E, Table 1) (Lyngwi, Koijam, Sharma & Joshi, 2013) based on their ethnomedicinal usages (Hynniewta & Kumar, 2008; Syiem et al. 2009). The taxonomic identities of the plants were confirmed with the help of the Herbarium Curator of the parent university. All samples were collected in sterile polythene bags and brought to the laboratory for the isolation of endophytic and epiphytic bacteria within 24h of collection.
Isolation of endophytic and epiphytic bacteria: Endophytic bacteria were isolated from the ethnomedicinal plants using methods of Hallmann, Quadt-Hallmann, Rodríguez-Kábana & Kloepper (1998), Strobel, Daisy, Castillo, & Harper (2004) and Long, Furuya, Kurose, Takeshita & Takanami (2003) with minor modifications. Plant samples (leaves, stems, roots, flowers, fruits) were washed thoroughly with tap water followed by sterile double distilled water and drained before immersion in 70% ethyl alcohol for 30s and sodium hypochlorite solution (2%) for 2min. The plant parts were then immersed in 70% ethyl alcohol for 30s (Barzanti et al. 2007) and then rinsed with sterile double distilled water. Each plant part was cut aseptically into 1cm long segments using a sterile blade under the laminar flow hood and allowed to dry. The cut surfaces of the segments were placed on Petri plates containing three different media viz., Tryptic Soya Agar (TSA), Nutrient Agar (NA) and Luria Bertani (LB) medium. The media was autoclaved at 121°C, 15psi for 20min and inoculation was carried out in the laminar flow to avoid contamination. Plates were then incubated at 32°C for 48h. Colonies with different morphology and pigmentation were randomly selected from each plate and streaked on fresh plates. Isolates were checked for purity by re-streaking on plates of the same medium and then transferred to nutrient agar slants where they were stored at 4°C. Simultaneously, the pure isolates were preserved in 20% glycerol at -20°C for further studies.
To analyze epiphytic microflora, plant parts (as above) were washed thoroughly with tap water followed by sterile double distilled water. No disinfectant was used in this case. The plant parts were dried under laminar flow hood and then placed on Petri plates containing the above mentioned three media. Plates were then incubated at 32°C for 48h. Colonies with different morphology and pigmentation were randomly selected from each plate and streaked on fresh plates and preserved as described above.
Molecular characterization of the isolates: Total genomic DNA was extracted using HiPurATM bacterial and yeast genomic DNA isolation kits (Himedia, India). PCR amplification and sequencing of 16S rRNA gene was carried out in a 25µL reaction mixture using general primers 27F 5’-AGAGTTTGATCCTGGCTGAG-3’and 1541R5’-AAGGAGGTGATCCAGCCGCA-3’ with the following conditions; denaturation at 94°C for 5min followed by 30 cycles at 94°C for 1min, annealing at 55°C for 1min, extension at 72°C for 2min, final step was carried out at 72°C for 5min and 4°C till infinity. The V6-V8 hypervariable region of bacteria-specific 16S rRNA gene was amplified with the primer pair 984GCF 5’-AACGCGAAGAACCTTAC-3’ and 1378R 5’-CGGTGTGTACAAGGCCCGGGAACG-3’ (Heuer, Krsek, Baker, Smalla & Wellington, 1997; Das, Royer & Leff, 2007) with the following conditions; denaturation at 94°C for 5min followed by 30 cycles at 94°C for 1min, annealing at 60°C for 1min, extension at 72°C for 2min, final step was carried out at 72°C for 5min and then 4°C till infinity using PCR Gene Amp 9700 (Applied Biosystems, USA). DNA template replaced with sterile water was used as negative control. The amplified 16S rRNA gene (approx. 1 400 base pairs) and V6-V8 region (approx. 400 base pairs) was then purified using QIAquick Gel Extraction Spin Kit (QIAGEN, Germany). Sequencing was performed using 20µL reaction mixture containing about 50ng DNA template (PCR product), 1pmol of sequencing primer, 3.5µL (5X) Big Dye buffer and 1µL Big Dye. This was carried out by denaturation step at 96°C for 5min, followed by 30 cycles at 96°C for 10s, 55°C (60°C for V6-V8 region) for 10s and final step at 96°C for 5min. Post reaction cleanup was carried out to remove unwanted matters including unincorporated dye terminators by using 125mM EDTA, 3M sodium acetate and 70% ethanol and then air dried at room temperature for 45min before keeping at -20°C. An amount of 10µL formamide was added and then denaturation step was carried out at 96°C for 2min, then immediately kept in ice before loading into the sequencer plate. Sequencing was performed using the Genetic Analyzer ABI 3130XL (Applied Biosystems, USA).
Phylogenetic analyses: Sequence similarities were determined using the BLAST program against the database of type strains with validly published prokaryotic names at the EzTaxon 2.1 server (Chun et al. 2007). Molecular Evolutionary Genetics Analysis software (MEGA version 4.0) (Tamura, Dudley, Nei, & Kumar, 2007) was used for phylogenetic analyses. The sequences of identified phylogenetic neighbours were aligned with the sequences of representative strains, using Clustal W inbuilt with MEGA 4. Neighbour-Joining method was employed to construct the phylogenetic tree with 1 000 bootstrap replications to assess nodal support in the tree.
Screening for Plant Growth Promoting (PGP) and Antagonistic Activitiesof the isolates
Phosphate Solubilization: Qualitative estimation of phosphate solubilization was performed by observing a halo zone around the bacterial colony grown in Pikovskaya medium (containing insoluble phosphate) (Kuklinsky-Sobral, Araújo, Mendes, & Geraldi, 2004) within 5-7days of incubation. Depending on the size of the halo zone, the respective isolates were scored positive. Quantitative estimation of phosphate solubilization was measured spectrophotometrically using the chlorostannous-reduced molybdophosphoric acid blue method (Jackson, 1973). After primary screening, the positive isolates were grown in 50mL of Pikovskaya broth in triplicate and incubated for 24, 48, 72 and 96h at 32°C. The uninoculated control was also kept under similar conditions. The cultures were harvested by centrifugation at 10 000rpm for 10min. To 1mL supernatant, 5mL of ammonium molybdate reagent (15g of ammonium molybdate in 400mL distilled water and 342mL of conc. HCl, total volume made up to 1 000mL) was added while shaking. A volume of 1mL of working solution of cholorostanous acid (0.5mL of stock solution was added to 65.5mL H2O to make final volume of 66mL) was added. Stock solution was made by dissolving 10g of SnCl2.2H2O in 25mL concentrated HCl. Total volume of reaction mixture was made to 25mL. O.D was measured at 660nm (Cecil CE7200, UK). Corresponding amount of soluble phosphorous was calculated from standard curve of potassium dihydrogen phosphate (KH2PO4; 0-10ppm).
Acid Phosphatase Assay: Acid phosphatase was estimated by adopting the modified assay of Tabatabai & Bremner (1969) at pH5.5 and pH4. The reaction mixture contained 1mL of the culture filtrate, 1mL of p-nitrophenyl- phosphate (pNPP) (0.05M), 4mL of 0.1M modified universal buffer (MUB) pH5.5/pH4 and incubated at 37ºC for 60min. The reaction was terminated by the addition of 4mL of NaOH (0.5M, pH10.8) and 1mL of CaCl2. All reactions were carried out in triplicate. The development of yellow colour was measured at 405nm. The amount of p-nitrophenol (pNP) released was quantified using the pNP standard. The enzyme activity was expressed in international unit (IU). One IU of phosphatase activity is the amount of enzyme required to release 1μmole of pNP per minute per mL of culture filtrate under the assay conditions.
Indole acetic acid (IAA) production: After screening the isolates for indole acetic acid (IAA) production, quantitative determination of free IAA like substances was performed. Isolates showing positive results were grown in LB (Luria Bertani) broth amended with 5mM tryptophan (Bric, Bostock, & Silverstone, 1991) and incubated at 32°C for five days in an environmental shaker at 120rpm. The broth was centrifuged at 10 000rpm for 15min at 4°C. A volume of 2mL of the supernatant was collected and 2-3 drops of o-phosphoric acid was added. The aliquots were shaken, and 4mL of Salkowski reagent (1mL of 0.5M FeCl3 and 50mL of 35% HClO4) (Gordon & Weber, 1951) was added and vortexed thoroughly. The samples were incubated at room temperature for 25min and their absorbance was read at 530nm (Cecil CE7200, UK). All reactions were carried out in triplicate. Auxin quantification value was recorded by extrapolating calibration curve made by using IAA as standard (1-50ppm).
PCR Screening for ACC gene: The gene encoding for 1-aminocyclopropane-1-carboxylic acid deaminase is ~800bp (Raddadi, Cherif, Boudabous & Daffonchio, 2008). The primers used were forward 5’-GTGAACCACCTGAATGTA-3’ and reverse 5’-AAACGAGATGATTTACTTGG-3’. Amplification was carried out using the following conditions: 94°C for 1min, 30 denaturation cycles at 94°C for 30s, annealing temperature at 52°C for 0.45s, primer extension at 72°C for 30s, followed by a final extension at 72°C for 5min.
Chitin degradation: A minimal salt medium containing colloidal chitin as a sole carbon and energy source was used to isolate chitinolytic bacteria. The medium consisted of Na2HPO4-6g; KH2PO4-3.0g; NH4Cl-1g; NaCl-0.5g; yeast extract-0.05g; colloidal chitin 1.0% (w/v) agar-15g and distilled water-1 000mL and incubated at 30ºC for 5days. Colloidal chitin was prepared by the method of Roberts & Selitrennikoff (1988) with minor modifications. 5g of chitin powder (HiMedia, India) was added into 60mL of concentrated HCl and left overnight at 4°C with vigorous stirring. The mixture was added to 500mL of ice-cold 95% ethanol with rapid stirring and kept overnight at room temperature (25°C). The precipitate was collected by centrifugation at 5 000xg for 20min at 4°C and was washed with sterile distilled water until the colloidal chitin became neutral (pH7.0).
Cellulose degradation: Cellulose-degrading ability of bacterial isolates was performed by streaking on the cellulose Congo- Red agar media with the following composition: KH2PO4-0.5g, MgSO4-0.25g, cellulose-2g, agar-15g, Congo Red-0.2g and gelatine-2g; distilled water-1 000mL and at pH 6.8-7.2. The use of Congo-Red as an indicator for cellulose degradation in an agar medium provides the basis for a rapid and sensitive screening test for cellulolytic bacteria (Lu, Wang, Yang, Wang & Nie, 2005). Colonies showing discoloration of Congo-Red were taken as positive cellulose-degrading bacterial colonies.
Pectin degradation: For screening of pectinase secreting bacteria, a modified medium was used consisting of: 5g pectin, 2g Ammonium sulphate; 6g Na2HPO4; 3g KH2PO4; 1g yeast extract; 20g agar (for 1 000mL) and at pH6. The petri plates were inoculated and incubated at 37oC for 24h and stained with 50mM iodine. Pectin degradation was evidenced by a clear zone around bacterial growth.
Nitrogen fixation: The ability of the isolates to fix atmospheric nitrogen was tested by growing bacteria in a nitrogen-free semisolid NFb malate medium (Okon, Albrecht, & Burris, 1977). After incubation for 3-5days at 32°C, the isolates showing growth at variable depth and change in colour under the surface medium were scored positive. Uninoculated NFb medium was kept as control.
Antagonistic activity: Bacterial isolates were grown in 100mL Nutrient Broth and incubated in a shaker incubator at 120rpm, 30°C for three days. The culture broths were centrifuged at 10 000rpm for 15min and the supernatant was filtered using autoclaved 0.22μm membrane filter paper. The filtrate was concentrated using Buchi rotavapor under reduced pressure at 40°C. The obtained aqueous extract was then tested for antimicrobial activity using agar well diffusion method against the plant pathogens, Fusarium oxysporum MTCC284 and Emericellanidulans var. nidulans MTCC1830. The test organisms were spread on the surface of Mueller-Hinton Agar (MHA, Difco) using a sterile swap stick. Wells of 3mm in diameter were cut into MHA plates by using a sterilized tip and 70μL of the extract was placed into each well. The plates were pre-incubated for two hours for diffusion and incubated aerobically overnight at 37°C. Zones of inhibition were measured and the mean recorded as described by Mukherjee, Balsubramanian, Saha, Pal & Saha (1995). Positive control was carried out under similar conditions by using nystatin and negative control was carried out using uninoculated media.
The experimental results were expressed as mean±standard deviation (SD) of three replicates (n=3). The results were processed using Origin 6.0.
Results
The ethnomedicinal plants of the study area (Table 1) were selected based on their medicinal usages (Table 2). A total of 70 plant associated bacteria were isolated from the 10 different ethnomedicinal plants, out of which 23 isolates were endophytic and 47 were epiphytes (Fig. 1). The study revealed significant differences in bacterial population not only between the epiphytic and endophytic microhabitats, but also amongst the host plants. Selected isolates were screened for PGP potential which revealed varying potentials (Table 3) for both endophytic and epiphytic bacteria. Plant associated bacteria showed remarkable ability to fix nitrogen, digest chitin, cellulose and pectin (Table 3, Fig. 2, Fig. 3) which is indicative of their ability to digest cell wall components. The isolates produced inhibitory metabolites that diffused into the agar medium and inhibited the growth of pathogens, resulting in the formation of inhibition zone (Fig. 3C, Fig. 3D). Among the endophytic isolates, B. siamensis C53 and B. subtilis cenB showed significant antagonistic activity while the epiphytic isolate showed no effect on the tested pathogens.
Plate screening method used for the detection of phosphate solubilizing isolates is indicated by the formation of clearing zones around the microbial colonies in the media containing insoluble mineral phosphate (). Quantitative test of the positive isolates with phosphate solubilization ability was then estimated. Phosphate solubilization was accompanied by the drop in pH of the culture medium. Maximum soluble phosphate was released at 72h of incubation by the majority of the isolates. The amount of soluble phosphorous was measured from the standard curve of potassium dihydrogen phosphate (R²=0.972, p<0.01). The amount of soluble phosphate released by endophytic isolate Lysinibacillus xylanilyticus C21 is 9.59ppm at 72h of incubation (Fig. 4A) with acid phosphatase activity of 0.047IU (Fig. 4B) whereas, epiphytic isolate Pantoea eucalypti CEN7E released the highest amount of soluble phosphate (9.69ppm at 72h of incubation) (Fig. 5A) with acid phosphatase activity of 0.042IU (Fig. 5B). Acid phosphatase activity of endophytes was estimated in the range of 0.015-0.0499IU while for epiphytes, the range was 0.0163-0.042IU. Acid phosphatase activity was higher at pH4 compared to pH5.5 (Fig. 4B and Fig. 5B).
The amount of IAA in the sample was measured from the standard IAA curve (R²=0.984, p<0.01). The quantity of IAA like substances detected (Fig. 3A) in endophytes was in the range of 1.80 to 10.72ppm and in epiphytes the range was 1.98 to 20.69ppm (Fig. 6). The epiphytic isolates, Raoultella ornithinolytica M11 produced the highest amount of IAA like substance (20.694ppm) followed by P. eucalypti CEN7E (12.61ppm) and the endophytic Pseudomonas palleroniana Y1 produced the least (10.72ppm). No detectable amount of IAA was observed in the isolates B. siamensis C53, B. methylotrophicus PotA and B. subtilis cenB (Fig. 6).
Rooted Neighbour-Joining Tree of the isolated endophytic and epiphytic bacteria with their nearest homologues of NCBI based on 16S rRNA gene sequences with 1 000 bootstrap replications for nodal support are represented in Fig. 7A and Fig. 7B. The scale bars indicate 0.02 change per nucleotide position for the 16S rRNA gene of the endophytic isolates and 0.05 change per nucleotide position of the epiphytic isolates. ACC deaminase gene of ~800bp was amplified for Bacillus sp. F21, Bacillus thuringiensis F41 and Bacillus mycoides M31 and their GenBank accession numbers are KC815532, KC815533 and KC815534 respectively. Nucleotide sequences of 16SrRNA and ACC deaminase encoding genes was deposited in NCBI.
Discussion
The results provided a preliminary understanding of plant growth promoting and antagonistic activities of associated endophytic and epiphytic bacteria with the selected ethnomedicinal plants. These plant associated bacteria are important determinants of ethnomedicinal plant community, as their presence effect the competition and coexistence of plants as well as soil nutrient dynamics (Rutherford et al. 2002). The present study revealed ethnomedicinal plants as the greatest potential resource for acquiring novel microorganisms and their products. Characterized endophytes and epiphytes in the present study revealed good plant promoting activities. Phosphorus is one of the major essential macronutrients for plant growth and development (Ehrlich, 1990). The conversion of insoluble phosphate (both organic and inorganic) compounds into a form accessible to the plant is an important trait in increasing plant yield (Rodríguez, Fraga, Gonzalez, & Bashan, 2006). The study revealed the release of soluble phosphate to be maximum at 72h of incubation by majority of the endophytic and epiphytic isolates. This is accompanied by the drop in pH value of the culture medium suggesting that organic acid secretion by the bacteria play a role in phosphate solubilizing activity (Rodríguez & Fraga, 1999). In the present study, both endophytic and epiphytic bacteria showed remarkable ability to release available phosphorus with acid phosphatase activity. Significant acid phosphatase activity was observed among the phosphate solubilizing isolates indicating a major role of acid phosphatase in this process.
A mode of action of some plant growth promoting microorganisms is the production of ACC deaminase, an enzyme that cleaves ACC into ammonia and a-ketobutyrate which are readily metabolized by bacteria (Klee, Hayford, Kretzmer, Barry, & Kishore, 1991). Three isolates of Bacillus species showed positive for ACC deaminase. The bacterium thus can act as a sink for ACC and lower ethylene level in plants, preventing the potentially deleterious consequences of high ethylene concentrations (Glick, Penrose, & Li, 1998). Production of ACC deaminase and IAA is an important and efficient way for plant growth promoting microorganisms to manipulate their plant hosts (Long, Schmidt, & Baldwin, 2008). IAA is a phytohormone involved in root initiation, cell division and cell enlargement (Salisbury, 1994) that enhances root growth and root length, resulting in greater root surface area and enables the plant to access more nutrients from the soil. The study revealedepiphytes as better IAA producers than endophytes.
The study revealed two endophytic isolates with antagonistic activity against the tested pathogens whereas none of the epiphytic isolates showed antagonistic activity. Endophytes are believed to carry out a resistance mechanism to overcome pathogenic invasion by producing secondary metabolites (Tan & Zou, 2001). Endophytes occupying the endosphere appear to have been selected by the host plant for their ability to resist the effects of plant defense products (Backman & Sikora, 2008) and a reason for the inability of epiphytes to colonize the internal host tissue. The study of this intriguing relationship between epiphytic and endophytic bacteria requires more attention, because the influence of epiphytes on their successful colonization as endophytes remains unclear and elusive. The plant growth promotion properties observed in the isolates of endophytes and epiphytes may provide tools for the conservation of the rapidly exploited medicinal plants prevalent in the studied subtropical forests and sustain their availability through cultivation in herbal gardens.
Acknowledgment
FMWN gratefully acknowledges University Grant Commission (UGC), Govt. of India for the fellowship and SRJ acknowledges DBT, Govt of India for financial support through a research grant.
References
Backman, P. A. & Sikora, R. A. (2008). Endophytes: An emerging tool for biological control. Biological Control, 46, 1-3. [ Links ]
Barzanti, R., Ozino, F., Bazzicalupo, M., Gabbrielli, R., Galardi, F., Gonnelli, C., & Mengoni, A. (2007). Isolation and characterization of endophytic bacteria from the nickel hyper accumulator plant Alyssum bertolonii. Microbial Ecology, 53, 306-316. [ Links ]
Bric, J. M., Bostock, R. M., & Silverstone, S. E. (1991). Rapid in situ assay for indoleacetic acid production by bacteria immobilized on nitrocellulose membrane. Applied and environmental Microbiology, 57, 535-538. [ Links ]
Chun, J., Lee, J. H., Jung, Y., Kim, M., Kim, S., Kim, B. K., & Lim, Y. W. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. International Journal of Systematic and Evolutionary Microbiology, 57, 2259-2261. [ Links ]
Das, M., Royer, T. V. & Leff, L. G. (2007). Diversity of fungi, bacteria, and actinomycetes on leaves decomposing in a stream. Applied and Environmental Microbiology, 73,756-767. [ Links ]
Ehrlich, H. L. (1990). Geomicrobiology (pp. 646). New York: Dekker. [ Links ]
Farnsworth, N. R., Akerele, O., Bingel, A. S., Soejarta, D. D., & Eno, Z. (1985). Medicinal plants in therapy. World Health Organ, 63, 965-981. [ Links ]
Glick, B. R., Penrose, D. M., & Li, J. (1998). A model for lowering plant ethylene concentration by plant growth promoting rhizobacteria. Journal of Theoretical Biology, 190,63-68. [ Links ]
Gordon, S. A. & Weber, R. P. (1951). Colorimetric estimation of indole-acetic acid. Plant Physiology, 26, 192-195. [ Links ]
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., & Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Canadian Journal of Microbiology, 43, 895-914. [ Links ]
Hallmann, J., Quadt-Hallmann, A., Rodríguez-Kábana, R., & Kloepper, J. W. (1998). Interactions between Meloidogyne incognita and endophytic bacteria in cotton and cucumber. Soil Biology & Biochemistry, 30, 925-937. [ Links ]
Hamayun, M., Khan, S. A., Khan, A. L., Rehman, G., Kim, Y. H., Iqbal, I., Hussain, J., Sohn, E. Y., & Lee, I. J. (2010). Gibberellins production and plant growth promotion by pure cultures of Cladosporium sp. MH-6 isolated from Cucumber (Cucumis sativus L.). Mycologia, 102, 989-995. [ Links ]
Heuer, H., Krsek, M., Baker, P., Smalla, K., & Wellington, E. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology, 63, 3233-3241. [ Links ]
Hynniewta, S. R. & Kumar, Y. (2008). Herbal remedies among the Khasi traditional healers and village folks in Meghalaya. Indian Journal of Traditional Knowledge, 7, 581-586. [ Links ]
Jackson, M. L. (1973). Soil Chemical Analysis (pp. 38-204). New Delhi: Prentice Hall of India Pvt Ltd. [ Links ]
Klee, H. J., Hayford, M. B., Kretzmer, K. A., Barry, G. F., & Kishore, G. M. (1991). Control of ethylene synthesis by expression of a bacterial enzymein transgenic tomato plants. Plant Cell, 3, 1187-1193. [ Links ]
Kobayashi, D. Y. & Columbo, J. D. (2000). Bacterial endophytes and their effects on plants and uses in agriculture. In C. W. Bacon & J. F. White (Eds.), Microbial Endophytes (pp. 199-236). New York: Marcel Dekker. [ Links ]
Kuklinsky-Sobral, J., Araújo, W. L., Mendes, R., & Geraldi, I. O. (2004). Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environmental Microbiology, 6, 1244-1251. [ Links ]
Long, H. H., Furuya, N., Kurose, D., Takeshita, M., & Takanami, Y. (2003). Isolation of endophytic bacteria from Solanum sp. and their antibacterial activity against plant pathogenic bacteria. Journal of the Faculty of Agriculture Kyushu University, 48, 21-28. [ Links ]
Long, H. H., Schmidt, D. D., & Baldwin, I. T. (2008). Native Bacterial Endophytes Promote Host Growth in a Species-Specific Manner; Phytohormone Manipulations Do Not Result in Common Growth Responses. PLoS ONE, 3, e2702. [ Links ]
Lu, W. J., Wang, H. T., Yang, S. J., Wang, Z. C., & Nie, Y. F. (2005). Isolation and characterization of mesophilic cellulose degrading bacteria from flower stalks vegetable waste co-composting system. Journal of General and Applied Microbiology, 51, 353-360. [ Links ]
Lyngwi, L. A., Koijam, K., Sharma, D., & Joshi, S. R. (2013). Cultivable bacterial diversity along the altitudinal zonation and vegetation range of tropical Eastern Himalaya. Revista de Biología Tropical, 61, 467-490. [ Links ]
Mukherjee, P. K., Balsubramanian, R., Saha, K., Pal, M., & Saha, B. P. (1995). Antibacterial efficiency of Nelumbo nucifera (Nymphaeaceae) rhizome extract. Indian drugs, 32,274-276. [ Links ]
Okon, Y., Albrecht, S. L., & Burris, R. H. (1977). Methods for growing Spirillum lipoferum and for counting it in pure culture and in association with plants. Applied and Environmental Microbiology, 33, 85-88. [ Links ]
Petrini, O. (1991). Fungal endophytes of tree leaves. In J. H. Andrews & S. S. Hirano (Eds.), Microbial Ecology of Leaves (pp. 179-197). New York: Springer Verlag. [ Links ]
Raddadi, N., Cherif, A., Boudabous, A., & Daffonchio, D. (2008). Screening of plant growth promoting traits of Bacillus thuringiensis. Annals of Microbiology, 58, 47-52. [ Links ]
Roberts, W. K. & Selitrennikoff, C. P. (1988). Plant and bacterial chitinases differ in antifungal activity. Journal of General Microbiology, 134, 169-176. [ Links ]
Rodríguez, H. & Fraga R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnology Advances, 17, 319-339. [ Links ]
Rodríguez, H., Fraga, R., Gonzalez, T., & Bashan, T. (2006). Genetics of phosphate solubilization and its potential applications for improving plant growth promoting bacteria. Plant Soil, 287, 15-21. [ Links ]
Rutherford, R. S., Van Antwerpen, T., Conlong, D. E., Keeping, M. G., McFarlane, S. A., & Vogel, J. L. (2002). Promoting plant health: Potential for the use of plant-associated micro-organisms in the biological control of pathogens and pests in sugarcane. Proceedings of the South African Sugar Technologists’ Association, 76, 289-300. [ Links ]
Salisbury, F. B. (1994). The role of plant hormones. In R. E. Wilkinson (Ed.), Plant-environment interactions (pp. 39-81). New York: Marcel Dekker. [ Links ]
Sekar, S. & Kandavel, D. (2010). Interaction of plant growth promoting rhizobacteria (pgpr) and endophytes with medicinal plants-new avenues for phytochemicals. Journal of Phytology, 2, 91-100. [ Links ]
Stierle, A., Strobel, G., & Stierle, D. (1993). Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of pacific yew (Taxus brevifolia). Science, 260, 214-216. [ Links ]
Strobel, G., Daisy, B., Castillo, U., & Harper, J. (2004). Natural products from endophytic microorganisms. Journal of Natural Products, 67, 257-268. [ Links ]
Syiem, D., Sharma, R., & Saio, V. (2009). In vitro study of the antioxidant potential of some traditionally used medicinal plants of North-East India and assessment of their total phenolic content. Pharmacologyonline, 3, 952-965. [ Links ]
Tabatabai, M. A. & Bremner, J. M. (1969). Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biology & Biochemistry, 1, 301-307. [ Links ]
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24,1596-1599. [ Links ]
Tan, R. X. & Zou, W. X. (2001). Endophytes: a rich source of functional metabolites. Natural Product Reports, 18, 448-459. [ Links ]
Barzanti, R., Ozino, F., Bazzicalupo, M., Gabbrielli, R., Galardi, F., Gonnelli, C., & Mengoni, A. (2007). Isolation and characterization of endophytic bacteria from the nickel hyper accumulator plant Alyssum bertolonii. Microbial Ecology, 53, 306-316. [ Links ]
Bric, J. M., Bostock, R. M., & Silverstone, S. E. (1991). Rapid in situ assay for indoleacetic acid production by bacteria immobilized on nitrocellulose membrane. Applied and environmental Microbiology, 57, 535-538. [ Links ]
Chun, J., Lee, J. H., Jung, Y., Kim, M., Kim, S., Kim, B. K., & Lim, Y. W. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. International Journal of Systematic and Evolutionary Microbiology, 57, 2259-2261. [ Links ]
Das, M., Royer, T. V. & Leff, L. G. (2007). Diversity of fungi, bacteria, and actinomycetes on leaves decomposing in a stream. Applied and Environmental Microbiology, 73,756-767. [ Links ]
Ehrlich, H. L. (1990). Geomicrobiology (pp. 646). New York: Dekker. [ Links ]
Farnsworth, N. R., Akerele, O., Bingel, A. S., Soejarta, D. D., & Eno, Z. (1985). Medicinal plants in therapy. World Health Organ, 63, 965-981. [ Links ]
Glick, B. R., Penrose, D. M., & Li, J. (1998). A model for lowering plant ethylene concentration by plant growth promoting rhizobacteria. Journal of Theoretical Biology, 190,63-68. [ Links ]
Gordon, S. A. & Weber, R. P. (1951). Colorimetric estimation of indole-acetic acid. Plant Physiology, 26, 192-195. [ Links ]
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., & Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Canadian Journal of Microbiology, 43, 895-914. [ Links ]
Hallmann, J., Quadt-Hallmann, A., Rodríguez-Kábana, R., & Kloepper, J. W. (1998). Interactions between Meloidogyne incognita and endophytic bacteria in cotton and cucumber. Soil Biology & Biochemistry, 30, 925-937. [ Links ]
Hamayun, M., Khan, S. A., Khan, A. L., Rehman, G., Kim, Y. H., Iqbal, I., Hussain, J., Sohn, E. Y., & Lee, I. J. (2010). Gibberellins production and plant growth promotion by pure cultures of Cladosporium sp. MH-6 isolated from Cucumber (Cucumis sativus L.). Mycologia, 102, 989-995. [ Links ]
Heuer, H., Krsek, M., Baker, P., Smalla, K., & Wellington, E. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology, 63, 3233-3241. [ Links ]
Hynniewta, S. R. & Kumar, Y. (2008). Herbal remedies among the Khasi traditional healers and village folks in Meghalaya. Indian Journal of Traditional Knowledge, 7, 581-586. [ Links ]
Jackson, M. L. (1973). Soil Chemical Analysis (pp. 38-204). New Delhi: Prentice Hall of India Pvt Ltd. [ Links ]
Klee, H. J., Hayford, M. B., Kretzmer, K. A., Barry, G. F., & Kishore, G. M. (1991). Control of ethylene synthesis by expression of a bacterial enzymein transgenic tomato plants. Plant Cell, 3, 1187-1193. [ Links ]
Kobayashi, D. Y. & Columbo, J. D. (2000). Bacterial endophytes and their effects on plants and uses in agriculture. In C. W. Bacon & J. F. White (Eds.), Microbial Endophytes (pp. 199-236). New York: Marcel Dekker. [ Links ]
Kuklinsky-Sobral, J., Araújo, W. L., Mendes, R., & Geraldi, I. O. (2004). Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environmental Microbiology, 6, 1244-1251. [ Links ]
Long, H. H., Furuya, N., Kurose, D., Takeshita, M., & Takanami, Y. (2003). Isolation of endophytic bacteria from Solanum sp. and their antibacterial activity against plant pathogenic bacteria. Journal of the Faculty of Agriculture Kyushu University, 48, 21-28. [ Links ]
Long, H. H., Schmidt, D. D., & Baldwin, I. T. (2008). Native Bacterial Endophytes Promote Host Growth in a Species-Specific Manner; Phytohormone Manipulations Do Not Result in Common Growth Responses. PLoS ONE, 3, e2702. [ Links ]
Lu, W. J., Wang, H. T., Yang, S. J., Wang, Z. C., & Nie, Y. F. (2005). Isolation and characterization of mesophilic cellulose degrading bacteria from flower stalks vegetable waste co-composting system. Journal of General and Applied Microbiology, 51, 353-360. [ Links ]
Lyngwi, L. A., Koijam, K., Sharma, D., & Joshi, S. R. (2013). Cultivable bacterial diversity along the altitudinal zonation and vegetation range of tropical Eastern Himalaya. Revista de Biología Tropical, 61, 467-490. [ Links ]
Mukherjee, P. K., Balsubramanian, R., Saha, K., Pal, M., & Saha, B. P. (1995). Antibacterial efficiency of Nelumbo nucifera (Nymphaeaceae) rhizome extract. Indian drugs, 32,274-276. [ Links ]
Okon, Y., Albrecht, S. L., & Burris, R. H. (1977). Methods for growing Spirillum lipoferum and for counting it in pure culture and in association with plants. Applied and Environmental Microbiology, 33, 85-88. [ Links ]
Petrini, O. (1991). Fungal endophytes of tree leaves. In J. H. Andrews & S. S. Hirano (Eds.), Microbial Ecology of Leaves (pp. 179-197). New York: Springer Verlag. [ Links ]
Raddadi, N., Cherif, A., Boudabous, A., & Daffonchio, D. (2008). Screening of plant growth promoting traits of Bacillus thuringiensis. Annals of Microbiology, 58, 47-52. [ Links ]
Roberts, W. K. & Selitrennikoff, C. P. (1988). Plant and bacterial chitinases differ in antifungal activity. Journal of General Microbiology, 134, 169-176. [ Links ]
Rodríguez, H. & Fraga R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnology Advances, 17, 319-339. [ Links ]
Rodríguez, H., Fraga, R., Gonzalez, T., & Bashan, T. (2006). Genetics of phosphate solubilization and its potential applications for improving plant growth promoting bacteria. Plant Soil, 287, 15-21. [ Links ]
Rutherford, R. S., Van Antwerpen, T., Conlong, D. E., Keeping, M. G., McFarlane, S. A., & Vogel, J. L. (2002). Promoting plant health: Potential for the use of plant-associated micro-organisms in the biological control of pathogens and pests in sugarcane. Proceedings of the South African Sugar Technologists’ Association, 76, 289-300. [ Links ]
Salisbury, F. B. (1994). The role of plant hormones. In R. E. Wilkinson (Ed.), Plant-environment interactions (pp. 39-81). New York: Marcel Dekker. [ Links ]
Sekar, S. & Kandavel, D. (2010). Interaction of plant growth promoting rhizobacteria (pgpr) and endophytes with medicinal plants-new avenues for phytochemicals. Journal of Phytology, 2, 91-100. [ Links ]
Stierle, A., Strobel, G., & Stierle, D. (1993). Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of pacific yew (Taxus brevifolia). Science, 260, 214-216. [ Links ]
Strobel, G., Daisy, B., Castillo, U., & Harper, J. (2004). Natural products from endophytic microorganisms. Journal of Natural Products, 67, 257-268. [ Links ]
Syiem, D., Sharma, R., & Saio, V. (2009). In vitro study of the antioxidant potential of some traditionally used medicinal plants of North-East India and assessment of their total phenolic content. Pharmacologyonline, 3, 952-965. [ Links ]
Tabatabai, M. A. & Bremner, J. M. (1969). Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biology & Biochemistry, 1, 301-307. [ Links ]
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24,1596-1599. [ Links ]
Tan, R. X. & Zou, W. X. (2001). Endophytes: a rich source of functional metabolites. Natural Product Reports, 18, 448-459. [ Links ]
1. Microbiology Laboratory, Department of Biotechnology and Bioinformatics, North-Eastern Hill University, Shillong-793022, India; fenella08@gmail.com
2. Microbiology Laboratory, Department of Biotechnology and Bioinformatics, North-Eastern Hill University, Shillong-793022, India srjoshi2006@yahoo.co.in
Received 15-X-2013. Corrected 30-IV-2014. Accepted 29-V-2014.