Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.62 n.2 San José Apr./Jun. 2014
Genetic variability of the Common Snook Centropomus undecimalis (Perciformes: Centropomidae) in connected marine and riverine environments
Variabilidad genética del robalo común Centropomus undecimalis (Perciformes: Centropomidae) en ambiente marino y ribereño interconectados
Variabilidad genética del robalo común Centropomus undecimalis (Perciformes: Centropomidae) en ambiente marino y ribereño interconectados
Ulises Hernández-Vidal1*, Julia Lesher-Gordillo2*, Wilfrido M. Contreras-Sánchez2 & Xavier Chiappa-Carrara3*
*Dirección para correspondencia:
Abstract
The Common Snook, Centropomus undecimalis, inhabits riverine and marine areas of Southern Gulf of Mexico, where it is subject to intense use and exploitation. It has been reported that the genetic identification of fish stocks constitutes a valuable tool for wild population management; nevertheless, there is no available information on the genetic identification on fish stocks of this species in the region. The aim of this study was to determine the genetic relationship between C. undecimalis captured in marine and freshwater environments of the Gulf of Mexico and the San Pedro River. For this, muscle tissue samples of 79 specimens were obtained from areas located more than 300km apart. The genotype of each individual was determined using seven microsatellite primer pairs. Five primers amplified efficiently presenting between six and 28 alleles per locus. High levels of heterozygosis were observed in samples from both environments. Deviation from HWE due to an excess of heterozygotes was observed. The values of genetic difference indicate an absence of population structure (FST=0.0075 and RST=0.016, p=0.051) and similarity in the allele frequencies, defined by Nei’s index (0.805). Data showed the existence of a high gene flow due to the number of migrants (Nm=18.7). Our results suggest that individuals living in these environments belong to the same genetic population. We suggest the development of management and protection plans for this fish species population in the wild. Rev. Biol. Trop. 62 (2): 627-636. Epub 2014 June 01.
Key words: genetic diversity, microsatellite, Common Snook, Centropomus undecimalis, Grijalva-Usumacinta fluvial system, Gulf of Mexico.
Resumen
El robalo común Centropomus undecimalis habita en áreas ribereñas y marinas del sur del Golfo de México donde es sujeto a explotación intensiva. Aunque la identificación de las poblaciones de peces representa una valiosa herramienta para el manejo de las poblaciones silvestres, no hay información disponible para identificar genéticamente las poblaciones de peces de esta especie en la región. El objetivo de este estudio fue determinar la relación genética entre C. undecimalis capturado en ambiente marino y dulceacuícola del Golfo de México y río San Pedro. Muestras de tejido muscular de 79 individuos fueron obtenidas en áreas separadas por más de 300km. El genotipo de cada individuo fue determinado usando siete pares de cebadores microsatélites. Cinco cebadores amplificaron eficientemente presentando entre seis y 28 alelos por locus. Altos niveles de heterocigosidad se observaron en las muestras de ambos ambientes. Se observó desviación del equilibrio HW debido a exceso de heterocigotos. Los valores de diferenciación genética indican ausencia de estructuración poblacional FST (0.0075) y RST (0.016, p=0.051) y similitud en las frecuencias alélicas definidas por el índice de Nei (0.805). Los datos mostraron elevado flujo genético debido al número de migrantes (Nm=18.7). Estos resultados sugieren que los individuos en estos ambientes provienen de la misma población genética. La información obtenida en este estudio, por lo tanto contribuirá con elementos que pueden ser considerados en el desarrollo de programas de manejo y protección de las poblaciones de peces silvestres.
Palabras clave: variabilidad genética, microsatélites, robalo común, Centropomus undecimalis, Sistema fluvial Grijalva-Usumacinta, Golfo de México
The Common Snook, Centropomus undecimalis, is a euryhaline species with migratory activity between marine, estuarine and fluvial environments throughout its life cycle. Its geo- graphic range is limited to the Atlantic coast of the American continent, extending from Florida, USA, to Brazil (McMichael, Peters, & Parsons, 1989; Tringali & Bert, 1996; Tringali, Bert, & Seyoum, 1999; Taylor, Whittington, Grier, & Crabtree, 2000). Reproduction of C. undecimalis in the Gulf of Mexico has been reported to occur in subtidal areas along the coast or within estuaries and coastal lagoons. Spawning takes place between April and September at salinities ranging from 28 to 35psu (Tucker, 1987; Tringali & Bert, 1996; Grier & Taylor, 1998).
C. undecimalis is the most economically valuable species captured in the Southern Gulf of Mexico; it represents one important target for artisanal and sport fisheries, along with other species of the genus Centropomus. It is mainly captured in the coastal areas and epicontinental tributaries of this region, where fisheries are related to its life cycle (Anonymous, 2006; Perera, Mendoza, Contreras, Huerta, & Pérez, 2011; Perera-García et al., 2013). Capture is strongly associated to migratory movements in freshwater ecosystems; while in the coastal zone, capture is linked to spawning events. These situations can promote stock depletion with potential detrimental effects on population conservation (Perera et al., 2011).
The Grijalva-Usumacinta fluvial system is the largest in Central America; it discharges into the Gulf of Mexico at the Campeche Bank area. One of the most important tributaries of this system is the San Pedro River -a fresh- water tributary-located near the border with Guatemala. This river connects with a wide wetland network (Castillo-Domínguez, Barba, Navarrete, Rodiles-Hernández, & Jiménez, 2011). Freshwater discharges to the coastal area of Tabasco vary according to the dry (Feb-Jun) and rainy (Jul-Jan) seasons. The coast has sandy beaches derived from sediment loads carried by rivers and shows an average salinity around 35psu but it reduces during the rainy season (Rosales-Hoz, Carranza-Edwards, Arias-Reynada, & Santiago-Pérez, 1992).
Juvenile and large adult snooks of both sexes can be found in riverine sites as far as 300km from the coast (where the known breeding areas are located) (Chávez, Mattheeuws, & Pérez, 1989; Perera, Mendoza & Páramo, 2008; Perera et al., 2011). Prodocimo et al. (2008) have shown that neighboring populations of Centropomid species do not differ over a wide geographical range, but several authors have demonstrated that genetic differences exist and, in some cases, these differences increase as geographic distance increases (Sandoval-Castellanos, Uribe, & Díaz, 2005; Diaz-Jaimes, Sandoval, & Uribe, 2007; Tringali et al., 2008). Whether C. undecimalis inhabiting environments hundreds of kilometers away, and as dissimilar as marine and freshwater are genetically related, has remained as an unsolved question.
Knowledge of a species genetic diversity has been used as a tool for developing management plans for fish stocks worldwide due to the fact that identification of a fishery unit is fundamental in the decision-making process for management strategies. The priority is to guarantee the sustainability of populations within their geographic range despite habitat alterations (Ward, 2000; Pritchard, Jones, & Cowley, 2007; Mehner, Pohlmann, Elkin, Monaghan, & Freyhof, 2009). Environmental differences found along the coastline play both an ecological and evolutionary role by favoring genetic differentiation (D´Anatro, Pereira, & Lessa, 2011) since the adaptive capacity of fish to withstand environmental changes determines the possibility of widening their distribution range and form new population units.
In this study we analyzed if C. undecimalis individuals form a single genetic unit even though they are present in two connected, but dissimilar and distant environments. This is important since Snooks are exploited with different strategies and intensities in each geographical location. Understanding this species genetic diversity and population structure can eventually translate into management and conservation strategies for one of the most important fishery resources in the Southern Gulf of Mexico (Anonymous, 2006).
Materials and methods
Study area: Two sites where selected considering a marine (MA) and a freshwater-riverine (FW) environment. The MA site was located off the coast of Tabasco (18°27.56’ -18°38.67’N; 92°42.57’-92°55.58’W). Monthly average salinities fluctuated between 24 and 35psu; yearly water temperature ranged between 24.0 and 29.4°C. The FW site was located on the San Pedro River’s main channel, one of the many tributaries of the Grijalva-Usu-macinta fluvial system. This sampling site is located near the Guatemala border, at 17°45.16’- 17°46.24’N; 91°12.75’-91°17.68’W. Water salinity values were between 0.10 and 0.90psu while water temperature oscillated between 22.6 and 32.3°C. The approximate distance between sampling sites was 320km (Fig. 1).
Sampling: A total of 79 specimens of both sexes were obtained from January to December 2010: 40 individuals from MA site weighing between 3.42 and 7.47kg, and 39 fish weighing between 0.5 and 10.75kg from the FW site. All fish were obtained on a monthly basis directly from gillnets used by local fishers in each sampling site, and geographical position was recorded (Fig. 1). Specimens were placed in iced water immediately after capture and sacrificed by a sharp blow on the head (DeTolla et al., 1995). Tissues from a minimum of three and a maximum of four fishes were processed each month. To obtain tissue samples, scales from the ventral area were removed and the excising area was treated with 70% ethylic alcohol. Skin was then removed, and a piece of muscle tissue (1-2g) was excised from the base of the pectoral fin. The muscle sample was placed in a sterilized plastic vial and preserved with ice during field activities and were subsequently frozen in the laboratory at -80°C until further analysis.
DNA extraction and PCR: DNA extraction from the samples was performed with a slight modification of the guanidine isothiocyanate technique (Tri reagent, Sigma Chemicals technical bulletin MB-205). This modification consists of additional steps at the end of the suggested protocol. After centrifugation to eliminate insoluble material, a volume of 1M-sodium acetate solution (10% of the sample volume) and 96% ethanol (200% of the sample volume) were added. The solutions were slowly mixed and allowed to stand for 5min. The DNA pellet was recovered by centrifugation at 14 000rpm during 10min at 4ºC. At the end, the DNA was dried and dissolved in ultrapure water. The DNA quality and quantity from each sample was determined with a Smartspec® (Bio-Rad) spectrophotometer. For further verification of DNA quality, electrophoresis was performed in Agarose gel at 0.5% with ethidium bromide, using TAE 1X as electrophoresis buffer. Electrophoresis was carried out for 90min at 90V and the extracted DNA was visualized in a UV transilluminator adapted with a system for the capture of digital images UVITEC. All DNA samples were treated with 6μL proteinase K (20mg/mL of distilled water) to optimize quality (Gamboa- Coronado, Mau-Inchaustegui, & Rodríguez- Cavallini, 2011).
Seven microsatellites designed for C. undecimalis (Cun01, Cun08, Cun09, Cun10A, Cun11, Cun18 and Cun22) were amplified using a slight modification of the PCR tech- nique described by Seyoum, Tringali, and Sullivan, 2005. Total volume of each PCR reaction was 20μL, consisting of 2μL of DNA (4ng/µL) and 18µL of master mix comprising 15µL of supermix (iQ Supermix, Bio Rad), 1µL of ultrapure sterile water and 1µL of each primer (forward and reverse). PCR reactions were performed using a thermocycler (MyCycler, Bio Rad) with the following amplification conditions: initial denaturation at 95°C for 5min, followed by seven cycles of 94°C for 30s, annealing temperature of 60°C for 1min with an increase gradient of 1°C per cycle, and an extension at 72°C for 1min, followed by 30 cycles at 94°C for 30s of denaturation, 55°C for 1min of annealing temperature, and an extension of 72°C for 1min, and lastly a final extension at 72°C for 10min, after the samples were maintained at 4°C. All PCR reactions were run in duplicates. To verify which micro- satellites amplified successfully, PCR products were visualized in a 2% agarose gel in 1X TAE buffer at 90V for 150min and stained with ethidium bromide. For allele size determination, automatic electrophoresis was performed using the Experion System (DNA 1K chip kit, Bio Rad).
The allele data from each specimen were captured in a matrix to analyze the allelic frequency. The Genepop software version 4.1.3 (Raymond & Rousset, 1995) was used to perform the Hardy-Weinberg equilibrium (HWE) global test using the Markov method combined with the exact test of Fisher (10 000 dememorizations, 50 batches and 5 000 iterations per batch) and the evaluation of the null hypothesis of heterozygote excess based on Markov’s method (Guo & Thompson, 1992). Genetic variability among environments was evaluated by means of Wright’s F statistics (1978); the statistics FIT, FST and FIS were calculated according to Weir and Cockerham (1984). Fisher’s method was used to evaluate genotypic linkage disequilibrium (10 000 dememorizations, 50 batches and 5 000 iterations per batch). The null allele frequency based on the EM algorithm developed by Dempster, Laird, and Rubin (1977) was obtained. The software Micro-Checker 2.2.3 (Van Oosterhout, Hutchinson, Wills, & Shipley, 2004) was used to estimate the most likely reason (presence of null alleles, evidence of allele dropout, or stuttering during PCR amplification), because genotype frequencies deviated significantly from HWE expectations. The program GenAlEX 6 (Peakall & Smouse, 2006) was used to obtain several parameters: observed heterozygosity (Ho) and expected heterozygosity (He), number of alleles per locus (Na), effective number of alleles per locus (Ne), polymorphism percentage per location, private alleles (Pa), null alleles frequency (Nf), gene flow by the number of migrants per generation (Nm), similarity degree by means of the genetic distance with Nei´s index (1978) per environment, R statistics of genetic differentiation (RIT, RST and RIS) according to Slatkin (1995) and the analysis of molecular variance (AMOVA), according to Excoffier, Smouse and Quattro (1992). In all cases, statistical differences were declared when p<0.05.
Results
Genetic variability: Genetic diversity obtained for five loci is shown in table 1. The total number of alleles per locus (Na) of organisms obtained in both environments ranged between six for the locus Cun09 and 28 for the locus Cun18 with an allele average of 21.1 (Table 1). Analysis per environment showed a lower allele average in FW (18.6) with a minimum of six alleles for locus Cun09 and a maximum of 28 for Cun18. In MA, allele average was 23.6 with a minimum of 20 for locus Cun22 and a maximum of 27 for Cun09.
High levels of heterozygosity were obtained in four loci from both environments. Average levels of Ho were generally above 0.50 for the loci Cun18 (0.830), Cun01 (0.884), Cun10A (0.834) and Cun09 (0.700) with the exception of the locus Cun22 with 0.442. Expected heterozygosity (He) average obtained for both environments were generally high for all the loci analyzed; the lowest being Cun09 with 0.854 and the highest Cun18 with 0.947 (Table 1). In every case, observed heterozygosity was lower than expected. The Markov-Fisher´s exact test for HWE on our data set showed deviations from the expected equilibrium (p<0.05). Micro-Checker software indicates high null allele frequency for some loci and no evidence of allele dropout or stuttering during PCR amplifications. The HWE deviation is consistent with the null hypothesis of excess of heterozygotes for the entire data set (p=0.31). Fish living in both environments were 100% polymorphic.
Locus Cun22 showed the highest null allele frequency. FW fish had a null allele frequency between 0.085 for the locus Cun09 and 0.301 for the Cun22. Data from MA fish were generally lower, and values ranged between 0 for Cun01 and 0.254 for Cun22 (Table 1). The Fisher test for the evaluation of genotype linkage disequilibrium suggests that the loci segregate individually with the exception of the pair Cun10A and Cun01 (p=0.03).
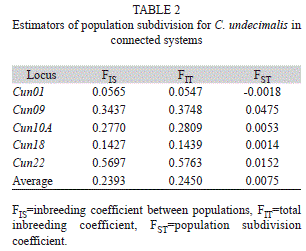
Genetic differentiation between both locations: Average values for Wright statistics were FST=0.0075 and FIS=0.2393 (Table 2). R-values were RST=0.016 (p>0.051) and RIS=0.173 (p<0.001). Average number of migrants per generation was Nm=18.7. Nei’s genetic distance showed a level of 0.805.
The AMOVA test showed decreasing values from within individuals (81%), among individuals (17%), and between environments (2%), results that provide further confirmation of the common origin of this population of high genetic diversity (Table 3).
Discussion
Genetic variation and similarity indices among sites showed that C. undecimalis tested from different environments belong to the same population. Results of FST and RST suggest a single population with high genetic flow and no subpopulations. This could mean a low differentiation condition in subpopulations (Balloux & Lugon-Moulin, 2002; Freeland, 2005). This is also evident in allelic frequency similarity as shown by Nei’s index and migrants per generation values. The estimated rate of migrants per generation (Nm=18.7) prevents divergence by genetic drift (Slatkin, 1994; Freeland, 2005). D´Anatro et al. (2011) came to the same conclusion with Micropogonias furnieri populations in Uruguay estuaries, where Nm=2.9. Low population differentiation levels are common in fish species that breed in the ocean due to mechanisms that ensure constant gene flow among distant areas (Ward, 2000).
Absence of genetic differentiation in the Common Snook suggests that the Grijalva-Usumacinta fluvial system’s environmental gradient is not a geographic barrier, likely allowing gene flow between marine and freshwater environments, over distances above 300km. Quite different is the case of Lutjanus synagris in coastal areas (Landinez-García, Ospina, Rodríguez, Arango, & Márquez, 2009), where habitat discontinuity due to environmental and geomorphological variation has been considered to drive population divergence. Divergence in marine populations of Centropomus viridis and C. medius in the Pacific Ocean coast increases with geographical distance. In this region, the absence or presence of water discharges into the coastal zone seems to play an important role in gene flow, because the specific conditions they generate are essential to maintain connectivity (Diaz-Jaimes et al., 2007).
In this study, the absence of genetic differentiation among environments, indicating no subpopulation in the Common Snook, is consistent with data suggesting absence of reproduction in the freshwater site reported by Perera et al. (2011). Additionally, Tucker (1987) and Taylor, Grier, and Whittington (1998) have suggested that spawning and larvae development occur in marine environments. This strengthens the hypotheses that different age and sex C. undecimalis in the San Pedro River are specimens from the marine population that moved to freshwater for food and shelter (Perera et al., 2008, 2011). The species’ great physiological capacity allows seasonal use of food and shelter resources. The same strategy has been reported in other Gulf of Mexico locations (Peterson & Gilmore,1991; Brennan, Walters, & Leber, 2008).
In contrast, physiologic tolerance mechanisms have been suggested as population divergence promoters in other euryhaline species. Although Micropogonias furnieri populations occur in different salinity habitats, they spawn in common coastal areas and juveniles apparently settle in their parents’ original environment, therefore favoring genetic differentiation (D´Anatro et al., 2011). Estuarine habitat colonization by Odontesthes argentinensis has contributed to its adaptive divergence expressed by specific breeding requirements that favor ecotype formation and population differentia- tion (Beheregaray & Sunnucks, 2001).
Genetic variation in individuals from marine and freshwater environments was high. Allele number and size were slightly different than the same lociones described by Seyoum et al. (2005) for C. undecimalis in Florida. Heterozygosity and polymorphism values were also high. Our results for He and Ho in most loci were consistent with Seyoum et al. (2005), except locus Cun22; He was 0.80-0.91 and Ho 0.74-0.91 whilst in this study values ranged from 0.76-0.94 and 0.60-0.96, respectively. In both cases, population genetic variation can be considered as high. High levels of genetic variation are common in marine fish due to the effect of environmental conditions on larvae dispersion as well as random mutation fixation processes (Tringali et al., 1999; Ward, 2000; Diaz-Jaimes et al., 2007; Landinez-García et al., 2009; Was, Gosling, & Hoarau, 2010). However, larval active movement ability in some species can hinder dispersion and favor population differentiation. Such is the case of Sebastes rastrelliger, a marine fish, whose larvae remain near their spawning area, thus reducing genetic exchange with other populations (Buonaccorsi et al., 2004).
Findings of this study allow us to assert that the observed variation is mainly generated by allelic frequencies in C. undecimalis within sites as opposed to between sites. Snook biological traits combined with oceanographic processes, could be involved with the high genetic variation registered in our study sites as well as in other localities. In coastal areas, common snook aggregations are a regular breeding behavior during the spawning season (Taylor et al., 1998; Perera et al., 2011), but the source of breeders in these aggregations is unknown. It is quite likely that specimens from both sites in this study exchange genetic material with other populations in Southern Gulf of Mexico. Furthermore, eggs and larvae can disperse over great distances with the strong currents caused by summer storms in the Gulf of Mexico (Salas, Monreal, & Aldeco, 1992; Expósito, Salas, Monreal, Salas, & Vázquez, 2009), that occur during this species’ spawning season (Roberts et al., 1999; Perera et al., 2008; 2011). Combination of spawning and early development with coastal currents circulation could play an important role in larval snook dispersion at a regional scale, such as could occur with Atlantic and Gulf of Mexico populations in Florida (Tringali et al., 2008). Dispersal associated with ocean circulation patterns has also been suggested for other Centropomids (Prodocimo et al., 2008); C. parallelus in coastal Brazil and C. viridis and C. medius of the Pacific coast in Mexico (Diaz-Jaimes et al., 2007). At a larger scale, however, ocean circulation patterns can act as geographical barriers for C. undecimalis, as suggested by Trigali and Highman (2007) based on differentiation between Atlantic and Caribbean populations. The same pattern was observed in Anguilla marmorata populations in the Indian and Pacific Oceans (Ishikawa, Tsukamoto, & Nishida, 2004).
Seemingly, deviation from the Hardy- Weinberg equilibrium model due to heterozygous excess is neither seemingly related to a large allele dropout nor stuttering during PCR amplification. The individual results for loci Cun22 and Cun09, that showed that they largely contribute to the genetic differentiation index (FST), inbreeding (FIS), high level of private alleles and null alleles frequency, should be taken with caution. The existence of selective processes for these loci is possible (Freeland, 2005), as well as faulty primer amplification due to base substitution or deletions in bonding sites for PCR (Was et al., 2010). Faulty amplification is an error that can favor null alleles and even amplification absence as with Cun08 and Cun11 (Was et al., 2010). Although there are no previous studies in the study sites that indicate a tendency to these results, their assessment is necessary, since amplification flaws can be an error source when interpreting genetic analysis (Trigali & Higham, 2007; Johnson & Banks, 2008; Landinez-García et al., 2009).
Snooks in freshwater and marine areas connected by the Grijalva-Usumacinta fluvial system can be considered a single fishery stock. This information is important to establish stock conservation programs that contribute maintaining migration routes between these systems. According to Perera et al. (2011) C. undecimalis population considered in this study seems to be under intensive extraction. This is important, since long term overexploitation could affect allelic frequency (Ward, 2000; Landinez-García et al., 2009) by gene flow reduction due to loss of connectivity between populations. This has been previously suggested for other Centropomus species of the Pacific coast in Mexico (Diaz-Jaimes et al., 2007).
Our results suggest that gene flow exists along different and distant environments where C. undecimalis inhabits, thus the null hypotheses that they form a single population is not rejected. Environmental differences, such as salinity ranging from 0.1-35ups, are not a barrier to gene flow; thus, for management purposes, individuals from both sites should be considered members of the same fishery stock. Therefore, to allow gene flow between these environments, we suggest implementing regulatory policies during the breeding season as well as freshwater habitat protection to maintain migratory routes and riparian feeding areas. New studies –focused on a wider geographical area and in the role of sea current on spatial-temporal variations –are required to identify possible genetic links in populations along the Gulf of Mexico coast. These studies will allow a better understanding of the Common Snook population dynamics and provide elements for sustainable management of this region’s important fishery resource.
Acknowledgments
This research is a component of the AquaFish Collaborative Research Support Program (CRSP), supported by the US Agency for Inter- national Development (USAID) award number CA/LWA No. EPP-A-00-06-0012-00 and by contributions from participating institutions. The AquaFish CRSP accession number is 1404. This study was partially supported by PFICA research program at UJAT.
References
Anonymus, (2006). Carta Nacional Pesquera. Secretaria de Agricultura, Ganadería, Pesca y Alimentación, México. Retrieved from http://www.inapesca.gob.mx [ Links ]
Balloux, F. & Lugon-Moulin, N. (2002). The estimation of population differentiation with microsatellite markers. Molecular Ecology, 11, 155-165. [ Links ]
Beheregaray, L. B. & Sunnucks, P. (2001). Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Molecular Ecology, 10, 2849-2866. [ Links ]
Brennan, N. P., Walters, C. J., & Leber, K. M. (2008). Manipulations of stocking magnitude: Addressing density dependence in a juvenile cohort of common snook (Centropomus undecimalis). Reviews in Fisheries Science, 16, 215-227. [ Links ]
Buonaccorsi, V. P., Westerman, M., Stannard, J., Kimbrell, C., Lynn, E., & Vetter, R. D. (2004). Molecular genetic structure suggests limited larval dispersal in grass rockfish, Sebastes rastrelliger. Marine Biology, 145, 779-88. [ Links ]
Castillo-Domínguez, A., Barba, M. E., Navarrete, A., Rodiles-Hernández, R., & Jiménez, B. (2011). Ictio-fauna de los humedales del río San Pedro, Balancán, Tabasco, México. Revista de Biología Tropical, 59, 693-708. [ Links ]
Castillo-Domínguez, A., Barba, M. E., Navarrete, A., Rodiles-Hernández, R., & Jiménez, B. (2011). Ictio-fauna de los humedales del río San Pedro, Balancán, Tabasco, México. Revista de Biología Tropical, 59, 693-708. [ Links ]
Chávez, L. M., Mattheeuws, O., & Pérez, M. H. (1989). Biología de los peces del río San Pedro en vista de determinar su potencial para la piscicultura. INREB- FUCID. Veracruz, México. [ Links ]
D´Anatro, A., Pereira, A. N., & Lessa, E. P. (2011). Genetic structure of the white croaker, Micropogonias furnieri Desmarest 1823 (Perciformes: Sciaenidae) along Uruguayan coasts: contrasting marine, estuarine, and lacustrine populations. Environmental Biology of Fishes, 91, 407-420. [ Links ]
Dempster, A. P., Laird, N. M., & Rubin, D. B. (1977). Maximum Likelihood from incomplete data via the EM algorithm (with discussion). Journal of the Royal Statistic Society, B(39), 1-38. [ Links ]
DeTolla, L. J., Srinivas, S., Whitaker, B. R., Andrews, C., Hecker, B., Kane, A. S., & Reimschuessel, R. (1995). Guidelines for the care and use of fish in research. ILAR Journal, 37(4), 159-173. [ Links ]
Díaz-Jaimes, P., Sandoval, E., & Uribe, M. (2007). Comparative population structure of three snook species (Teleostei: Centropomidae) from the eastern central Pacific. Ichthyological Research, 54, 380-387. [ Links ]
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491. [ Links ]
Expósito, G., Salas, D. A., Monreal, M. A., Salas, D., & Vázquez, F. (2009). Inertial currents in the southern Gulf of Mexico. Ciencias Marinas, 35, 287-296. [ Links ]
Freeland, J. (2005). Molecular Ecology. England: Wiley and Sons. [ Links ]
Gamboa-Coronado, M., Mau-Inchaustegui, S., & Rodríguez-Cavallini, E. (2011). Caracterización molecular y resistencia antimicrobiana de aislamientos de Clostridium perfringens de diferentes orígenes en Costa Rica. Revista de Biología Tropical, 59, 1479-1485. [ Links ]
Grier, H. J. & Taylor, R. G. (1998). Testicular maturation and regression in the common snook. Journal of Fish Biology, 53, 521-542. [ Links ]
Guo, S. W. & Thompson, E. A. (1992). Performing the exact test for Hardy-Weinberg proportion for multiple alleles. Biometrics, 48, 361-372. [ Links ]
Ishikawa, S., Tsukamoto, K., & Nishida, M. (2004). Genetic evidence for multiple geographic populations of the giant mottled eel Anguilla marmorata in the Pacific and Indian oceans. Ichthyological Research, 51, 343-353. [ Links ]
Johnson, M. A. & Banks, M. A. (2008). Genetic structure, migration, and patterns of allelic richness among coho salmon (Oncorhynchus kisutch) populations of the Oregon coast. Canadian Journal of Fisheries and Aquatic Sciences, 65, 1274-1285. [ Links ]
Landínez-García, R. M., Ospina, S. P., Rodríguez, D. J., Arango, R., & Márquez, E. (2009). Genetic analysis of Lutjanus synagris populations in the Colombian Caribbean. Ciencias Marinas, 35, 321-331. [ Links ]
McMichael, R. H., Peters, K. M., Jr., & Parsons, G. R. (1989). Early life history of the snook, Centropomus undecimalis, in Tampa Bay, Florida. North East Gulf Sciences, 10, 113-126. [ Links ]
Mehner, T., Pohlmann, K., Elkin, C., Monaghan, M. T. ,& Freyhof, J. (2009). Genetic mixing from enhancement stocking in commercially exploited ven- dace populations. Journal of Applied Ecology, 46, 1340-1349. [ Links ]
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590. [ Links ]
Peakall, R. & Smouse, P. E. (2006). GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295. [ Links ]
Perera, M. A., Mendoza, M., Contreras, W. M., Huerta, M., & Pérez, E. (2011). Reproductive biology of common snook Centropomus undecimalis (Perciformes: Centropomidae) in two tropical habitats. Revista de Biología Tropical, 59, 669-681. [ Links ]
Perera, M. A., Mendoza, M., & Páramo, S. (2008). Dinámica reproductiva y poblacional del robalo, Centropomus undecimalis (Perciformes: Centropomidae), en barra San Pedro, Centla, México. Universidad y Ciencia, 24(1), 49-59. [ Links ]
Perera-García, M., Mendoza-Carranza, M., Contreras-Sánchez, W., Ferrara, A., Huerta-Ortiz, M., & Hernández-Gómez, R. (2013). Comparative age and growth of common snook Centropomus undecimalis (Pisces: Centropomidae) from coastal and riverine areas in Southern Mexico. Revista de Biología Tropical, 61, 807-819. [ Links ]
Peterson, M. S. & Gilmore, R. G. (1991). Eco-physiology of juvenile snook Centropomus undecimalis (Bloch): life-history implications. Bulletin of Marine Science, 48(1), 46-57. [ Links ]
Pritchard, V. L., Jones, K., & Cowley, D. E. (2007). Estimation of introgression in cutthroat trout populations using microsatellites. Conservation Genetics, 8, 1311-1329. [ Links ]
Prodocimo, V., Tscha, M. K., Pie, M. R., Oliveira, J., Ostrensky, A., & Boerger, W. A. (2008). Lack of genetic differentiation in the fat snook Centropomus parallelus (Teleostei: Centropomidae) along the Brazilian coast. Journal of Fish Biology, 73, 2075-2082. [ Links ]
Raymond, M. & Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248-249. [ Links ]
Roberts, S. B., Jackson, L. F., King, W., Taylor, R. G., Grier, H. J., & Sullivan, C. V. (1999). Annual Reproductive Cycle of the Common Snook: Endocrine Correlates of Maturation. Transactions of the American Fisheries Society, 128, 436-445. [ Links ]
Rosales-Hoz, L., Carranza-Edwards, A., Arias-Reynada, S., & Santiago-Pérez, S. (1992). Distribución de metales en sedimentos recientes del sureste del golfo de México. Anales del Instituto de Ciencias del Mar y Limnología. http://biblioweb.tic.unam.mx/cienciasdelmar/instituto/1992-2/articulo408.html [ Links ]
Salas, L. A., Monreal, M. A., & Aldeco, J. (1992). Periodos característicos en las oscilaciones de parámetros meteorológicos en Cayo Arcas, México. Atmósfera, 5, 193-205. [ Links ]
Sandoval-Castellanos, E., Uribe, M., & Díaz, P. (2005). Diferenciación genética poblacional en robalos (Pisces:Centropomidae) del pacífico mexicano. Revista Internacional de Contaminación Ambiental, 21(1), 35-41. [ Links ]
Seyoum, S., Tringali, M. D., & Sullivan, J. (2005). Isolation and characterization of 27 polymorphic microsatellite loci for the common snook Centropomus undecimalis. Molecular Ecology Notes, 5, 192-194. [ Links ]
Slatkin, M. (1994). Gene flow and population structure. In L. A. Real (Ed.). Ecological Genetics (pp. 3-17). Nueva Jersey, USA: Princeton University. [ Links ]
Slatkin, M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics, 139, 457-462. [ Links ]
Taylor, R. G., Grier, H. J., & Whittington, J. A. (1998). Spawning rhythms of common snook in Florida. Journal of Fish Biology, 53, 502-520. [ Links ]
Taylor, R. G., Whittington, J. A., Grier, H. J., & Crabtree, R. E. (2000). Age, growth and protandric sex reversal in common snook Centropomus undecimalis, from the east and west coasts of Florida. Fishery Bulletin, 98, 612-624. [ Links ]
Tringali, M. D. & Bert, T. M. (1996). The genetic stock structure in common snook (Centropomus undecimalis). Canadian Journal of Fisheries and Aquatic Sciences, 53, 974-984. [ Links ]
Tringali, M. D. & Higham, M. (2007). Isolation by distance gene flow among Vermilion snapper (Rhomboplites aurorubens Cuvier, 1829) from the Gulf of Mexico and Southeastern United States. Gulf of Mexico Science, 25(1), 2-14. [ Links ]
Tringali, M. D., Bert, T. M., & Seyoum, S. (1999). Genetic identification of centropomine fishes. Transactions of the American Fisheries Society, 1(8), 446-458. [ Links ]
Tringali, M. D., Seyoum, S., Wallace, E. M., Higham, M., Taylor, R. G., Trotter, A. A., & Whittington, J. A. (2008). Limits to the use of contemporary genetic analyses in delineating biological populations for restocking and stock enhancement. Reviews on Fisheries Sciences, 16(1), 111-116. [ Links ]
Tucker, J. W. (1987). Snook and tarpon snook culture and preliminary evaluation for commercial farming. The Progressive Fish-Culturist, 49, 49-57. [ Links ]
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P., & Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology, 4, 535-538. [ Links ]
Ward, R. D. (2000). Genetics in fisheries management. Hydrobiologia, 420, 191-201. [ Links ]
Was, A., Gosling, E., & Hoarau, G. (2010). Microsatellite analysis of plaice (Pleuronectes platessa L.) in the NE Atlantic: weak genetic structuring in a milieu of high gene flow. Marine Biology, 157, 447-462. [ Links ]
Weir, B. S. & Cockerham, C. C. (1984). Estimating F statistics for the analysis of population structure. Evolution, 38, 43-44. [ Links ]
Balloux, F. & Lugon-Moulin, N. (2002). The estimation of population differentiation with microsatellite markers. Molecular Ecology, 11, 155-165. [ Links ]
Beheregaray, L. B. & Sunnucks, P. (2001). Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Molecular Ecology, 10, 2849-2866. [ Links ]
Brennan, N. P., Walters, C. J., & Leber, K. M. (2008). Manipulations of stocking magnitude: Addressing density dependence in a juvenile cohort of common snook (Centropomus undecimalis). Reviews in Fisheries Science, 16, 215-227. [ Links ]
Buonaccorsi, V. P., Westerman, M., Stannard, J., Kimbrell, C., Lynn, E., & Vetter, R. D. (2004). Molecular genetic structure suggests limited larval dispersal in grass rockfish, Sebastes rastrelliger. Marine Biology, 145, 779-88. [ Links ]
Castillo-Domínguez, A., Barba, M. E., Navarrete, A., Rodiles-Hernández, R., & Jiménez, B. (2011). Ictio-fauna de los humedales del río San Pedro, Balancán, Tabasco, México. Revista de Biología Tropical, 59, 693-708. [ Links ]
Castillo-Domínguez, A., Barba, M. E., Navarrete, A., Rodiles-Hernández, R., & Jiménez, B. (2011). Ictio-fauna de los humedales del río San Pedro, Balancán, Tabasco, México. Revista de Biología Tropical, 59, 693-708. [ Links ]
Chávez, L. M., Mattheeuws, O., & Pérez, M. H. (1989). Biología de los peces del río San Pedro en vista de determinar su potencial para la piscicultura. INREB- FUCID. Veracruz, México. [ Links ]
D´Anatro, A., Pereira, A. N., & Lessa, E. P. (2011). Genetic structure of the white croaker, Micropogonias furnieri Desmarest 1823 (Perciformes: Sciaenidae) along Uruguayan coasts: contrasting marine, estuarine, and lacustrine populations. Environmental Biology of Fishes, 91, 407-420. [ Links ]
Dempster, A. P., Laird, N. M., & Rubin, D. B. (1977). Maximum Likelihood from incomplete data via the EM algorithm (with discussion). Journal of the Royal Statistic Society, B(39), 1-38. [ Links ]
DeTolla, L. J., Srinivas, S., Whitaker, B. R., Andrews, C., Hecker, B., Kane, A. S., & Reimschuessel, R. (1995). Guidelines for the care and use of fish in research. ILAR Journal, 37(4), 159-173. [ Links ]
Díaz-Jaimes, P., Sandoval, E., & Uribe, M. (2007). Comparative population structure of three snook species (Teleostei: Centropomidae) from the eastern central Pacific. Ichthyological Research, 54, 380-387. [ Links ]
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491. [ Links ]
Expósito, G., Salas, D. A., Monreal, M. A., Salas, D., & Vázquez, F. (2009). Inertial currents in the southern Gulf of Mexico. Ciencias Marinas, 35, 287-296. [ Links ]
Freeland, J. (2005). Molecular Ecology. England: Wiley and Sons. [ Links ]
Gamboa-Coronado, M., Mau-Inchaustegui, S., & Rodríguez-Cavallini, E. (2011). Caracterización molecular y resistencia antimicrobiana de aislamientos de Clostridium perfringens de diferentes orígenes en Costa Rica. Revista de Biología Tropical, 59, 1479-1485. [ Links ]
Grier, H. J. & Taylor, R. G. (1998). Testicular maturation and regression in the common snook. Journal of Fish Biology, 53, 521-542. [ Links ]
Guo, S. W. & Thompson, E. A. (1992). Performing the exact test for Hardy-Weinberg proportion for multiple alleles. Biometrics, 48, 361-372. [ Links ]
Ishikawa, S., Tsukamoto, K., & Nishida, M. (2004). Genetic evidence for multiple geographic populations of the giant mottled eel Anguilla marmorata in the Pacific and Indian oceans. Ichthyological Research, 51, 343-353. [ Links ]
Johnson, M. A. & Banks, M. A. (2008). Genetic structure, migration, and patterns of allelic richness among coho salmon (Oncorhynchus kisutch) populations of the Oregon coast. Canadian Journal of Fisheries and Aquatic Sciences, 65, 1274-1285. [ Links ]
Landínez-García, R. M., Ospina, S. P., Rodríguez, D. J., Arango, R., & Márquez, E. (2009). Genetic analysis of Lutjanus synagris populations in the Colombian Caribbean. Ciencias Marinas, 35, 321-331. [ Links ]
McMichael, R. H., Peters, K. M., Jr., & Parsons, G. R. (1989). Early life history of the snook, Centropomus undecimalis, in Tampa Bay, Florida. North East Gulf Sciences, 10, 113-126. [ Links ]
Mehner, T., Pohlmann, K., Elkin, C., Monaghan, M. T. ,& Freyhof, J. (2009). Genetic mixing from enhancement stocking in commercially exploited ven- dace populations. Journal of Applied Ecology, 46, 1340-1349. [ Links ]
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590. [ Links ]
Peakall, R. & Smouse, P. E. (2006). GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295. [ Links ]
Perera, M. A., Mendoza, M., Contreras, W. M., Huerta, M., & Pérez, E. (2011). Reproductive biology of common snook Centropomus undecimalis (Perciformes: Centropomidae) in two tropical habitats. Revista de Biología Tropical, 59, 669-681. [ Links ]
Perera, M. A., Mendoza, M., & Páramo, S. (2008). Dinámica reproductiva y poblacional del robalo, Centropomus undecimalis (Perciformes: Centropomidae), en barra San Pedro, Centla, México. Universidad y Ciencia, 24(1), 49-59. [ Links ]
Perera-García, M., Mendoza-Carranza, M., Contreras-Sánchez, W., Ferrara, A., Huerta-Ortiz, M., & Hernández-Gómez, R. (2013). Comparative age and growth of common snook Centropomus undecimalis (Pisces: Centropomidae) from coastal and riverine areas in Southern Mexico. Revista de Biología Tropical, 61, 807-819. [ Links ]
Peterson, M. S. & Gilmore, R. G. (1991). Eco-physiology of juvenile snook Centropomus undecimalis (Bloch): life-history implications. Bulletin of Marine Science, 48(1), 46-57. [ Links ]
Pritchard, V. L., Jones, K., & Cowley, D. E. (2007). Estimation of introgression in cutthroat trout populations using microsatellites. Conservation Genetics, 8, 1311-1329. [ Links ]
Prodocimo, V., Tscha, M. K., Pie, M. R., Oliveira, J., Ostrensky, A., & Boerger, W. A. (2008). Lack of genetic differentiation in the fat snook Centropomus parallelus (Teleostei: Centropomidae) along the Brazilian coast. Journal of Fish Biology, 73, 2075-2082. [ Links ]
Raymond, M. & Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248-249. [ Links ]
Roberts, S. B., Jackson, L. F., King, W., Taylor, R. G., Grier, H. J., & Sullivan, C. V. (1999). Annual Reproductive Cycle of the Common Snook: Endocrine Correlates of Maturation. Transactions of the American Fisheries Society, 128, 436-445. [ Links ]
Rosales-Hoz, L., Carranza-Edwards, A., Arias-Reynada, S., & Santiago-Pérez, S. (1992). Distribución de metales en sedimentos recientes del sureste del golfo de México. Anales del Instituto de Ciencias del Mar y Limnología. http://biblioweb.tic.unam.mx/cienciasdelmar/instituto/1992-2/articulo408.html [ Links ]
Salas, L. A., Monreal, M. A., & Aldeco, J. (1992). Periodos característicos en las oscilaciones de parámetros meteorológicos en Cayo Arcas, México. Atmósfera, 5, 193-205. [ Links ]
Sandoval-Castellanos, E., Uribe, M., & Díaz, P. (2005). Diferenciación genética poblacional en robalos (Pisces:Centropomidae) del pacífico mexicano. Revista Internacional de Contaminación Ambiental, 21(1), 35-41. [ Links ]
Seyoum, S., Tringali, M. D., & Sullivan, J. (2005). Isolation and characterization of 27 polymorphic microsatellite loci for the common snook Centropomus undecimalis. Molecular Ecology Notes, 5, 192-194. [ Links ]
Slatkin, M. (1994). Gene flow and population structure. In L. A. Real (Ed.). Ecological Genetics (pp. 3-17). Nueva Jersey, USA: Princeton University. [ Links ]
Slatkin, M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics, 139, 457-462. [ Links ]
Taylor, R. G., Grier, H. J., & Whittington, J. A. (1998). Spawning rhythms of common snook in Florida. Journal of Fish Biology, 53, 502-520. [ Links ]
Taylor, R. G., Whittington, J. A., Grier, H. J., & Crabtree, R. E. (2000). Age, growth and protandric sex reversal in common snook Centropomus undecimalis, from the east and west coasts of Florida. Fishery Bulletin, 98, 612-624. [ Links ]
Tringali, M. D. & Bert, T. M. (1996). The genetic stock structure in common snook (Centropomus undecimalis). Canadian Journal of Fisheries and Aquatic Sciences, 53, 974-984. [ Links ]
Tringali, M. D. & Higham, M. (2007). Isolation by distance gene flow among Vermilion snapper (Rhomboplites aurorubens Cuvier, 1829) from the Gulf of Mexico and Southeastern United States. Gulf of Mexico Science, 25(1), 2-14. [ Links ]
Tringali, M. D., Bert, T. M., & Seyoum, S. (1999). Genetic identification of centropomine fishes. Transactions of the American Fisheries Society, 1(8), 446-458. [ Links ]
Tringali, M. D., Seyoum, S., Wallace, E. M., Higham, M., Taylor, R. G., Trotter, A. A., & Whittington, J. A. (2008). Limits to the use of contemporary genetic analyses in delineating biological populations for restocking and stock enhancement. Reviews on Fisheries Sciences, 16(1), 111-116. [ Links ]
Tucker, J. W. (1987). Snook and tarpon snook culture and preliminary evaluation for commercial farming. The Progressive Fish-Culturist, 49, 49-57. [ Links ]
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P., & Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology, 4, 535-538. [ Links ]
Ward, R. D. (2000). Genetics in fisheries management. Hydrobiologia, 420, 191-201. [ Links ]
Was, A., Gosling, E., & Hoarau, G. (2010). Microsatellite analysis of plaice (Pleuronectes platessa L.) in the NE Atlantic: weak genetic structuring in a milieu of high gene flow. Marine Biology, 157, 447-462. [ Links ]
Weir, B. S. & Cockerham, C. C. (1984). Estimating F statistics for the analysis of population structure. Evolution, 38, 43-44. [ Links ]
*Correspondencia a:
Ulises Hernández-Vidal:Posgrado en Ciencias del Mar y Limnología, sede Unidad Académica Sisal, Universidad Nacional Autónoma de México. Puerto de Abrigo S/N, Sisal, Yucatán C.P. 97355; uliseshv44@hotmail.com. Correspondence.
Julia Lesher-Gordillo: División Académica de Ciencias Biológicas, Universidad Juárez Autónoma de Tabasco. Carretera Villahermosa-Cárdenas km 0.5, Villahermosa, Tabasco C.P. 86280; lesher23@yahoo.com
Wilfrido M. Contreras-Sánchez: División Académica de Ciencias Biológicas, Universidad Juárez Autónoma de Tabasco. Carretera Villahermosa-Cárdenas km 0.5, Villahermosa, Tabasco C.P. 86280; contrerw@hotmail.com
Xavier Chiappa-Carrara: Unidad Multidisciplinaria de Docencia e Investigación, Unidad Académica Sisal, Universidad Nacional Autónoma de México. Puerto de Abrigo S/N, Sisal, Yucatán C.P. 97355; xcc@ciencias.unam.mx
1. Posgrado en Ciencias del Mar y Limnología, sede Unidad Académica Sisal, Universidad Nacional Autónoma de México. Puerto de Abrigo S/N, Sisal, Yucatán C.P. 97355; uliseshv44@hotmail.com. Correspondence.
2. División Académica de Ciencias Biológicas, Universidad Juárez Autónoma de Tabasco. Carretera Villahermosa-Cárdenas km 0.5, Villahermosa, Tabasco C.P. 86280; lesher23@yahoo.com, contrerw@hotmail.com
3. Unidad Multidisciplinaria de Docencia e Investigación, Unidad Académica Sisal, Universidad Nacional Autónoma de México. Puerto de Abrigo S/N, Sisal, Yucatán C.P. 97355; xcc@ciencias.unam.mx
Received 05-IV-2013. Corrected 20-X-2013. Accepted 28-XI-2013.