Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.52 n.3 San José Sep. 2004
The STR polymorphism (AAAAT)n within the intron 1 of the tumor
protein 53 (TP53) locus in 17 populations of different ethnic groups
of Africa, America, Asia and Europe
Jorge Azofeifa1 *, Meinhard Hahn2 , Edward Ruiz1 , Lars Hummerich2 , Ana I. Morales3, Gerardo Jiménez4 & Ramiro Barrantes1
1 Escuela de Biología and Instituto de Investigaciones en Salud (INISA), Universidad de Costa Rica
2 Institut für Biochemie, Justus-Liebig-Universität Giessen, Germany (Present address: Deutsches Krebsforschungszentrum, Abteilung Molekulare Genetik (H0700), Heidelberg, Germany)
3 Organismo de Investigaciones Judiciales, Costa Rica
4 Centro de Investigaciones en Hemoglobinas Anormales y Trastornos Afines (CIHATA), Universidad de Costa Rica
* Correspondence to: Jorge Azofeifa, Instituto de Investigaciones en Salud (INISA), Universidad de Costa Rica, Ciudad Universitaria "Rodrigo Facio", San José, Costa Rica, Central America. Tel.: (+506) 224 36 68, Fax: (+506) 207 51 30; azofeifa@biologia.ucr.ac.cr
Abstract
The STR (AAAAT)n within intron 1 of the TP53 locus was screened in 17 populations from 3 main ethnic groups: Europeans, Asiatics, and Africans, and from the hybrid population of Costa Rica (1968 samples). Three alleles, 126/7 (bp/copies of the repeat), 131/8 and 136/9 were the most prevalent in all populations. Other alleles rarely reached frequencies of 10% or higher. Observed heterozygosities ranged between 0.351 and 0.829. Patterns of diversity fit well with both the geographic origin of the samples and the history of the populations screened. A statistical test suggests that single-step mutational events have been the main mechanism producing new alleles at this locus. Fixation indexes (RST ) for this marker showed an effect of population subdivision on divergence only within the Asiatic group; they were insensitive at the level of major ethnic groups as well as within Africans and within Europeans. Rev. Biol. Trop. 52(3): 645-657. Epub 2004 Dic 15.
Key words: population genetics, microsatellite polymorphism, Europeans, Asiatics, sub-Saharan Africans.
Palabras clave: genética de poblaciones, microsatélites, polimorfismos, europeos, asiáticos, africanos del sub-Sahara.
Received 13-VIII-2003. Corrected 15-X-2003. Accepted 02-XI-2003.
The tumor protein 53 locus (TP53) (MIM No.191170, GDB ID 120445) maps to 17p13.1 (Isobe et al. 1986). The gene spans 12.5 kb, is segmented in 11 exons, the first of which is not translated, and codes for a 43.5 kDa protein (p53) involved in cellular responses to genetic damage as mediator in DNA repair, in growth arrest at the G1-S checkpoint of the cell cycle, or as an inductor of apoptosis (Hesketh 1997, Janus et al. 1999). Although several polymorphic regions have been described within the locus, population genetic studies are not abundant. Exceptional are the works of two groups which have studied the haplotypic variation defined by the presence/absence of a 16bp duplication within intron 3, of a BstU1 restriction site within exon 4 and of an MspI restriction site within intron 6 in Northern Europeans, Chinese and Africans (Själander et al. 1995, 1996), and in Amerindians, Blacks and European descendants from Brazil (Gaspar et al. 2001, 2002). On the other hand, several mutations have been directly implicated in approximately 50% of all human cancers (Janus et al. 1999) -for an extensive review see OMIM. In this association, Ridanpää et al. (1995) have studied samples of archival lung cancer tissue using a pentanucleotide (AAAAT)n within the 6.1 kb-long intron 1 of the locus, a polymorphism first reported by Futreal et al. (1991) and further characterized by Hahn et al. (1995). The methodological improvements developed by Hahn et al. (1993) facilitated the screening of large samples. Hence, population data of this polymorphism have been reported for two samples, one from Germany (Hahn et al. 1995) and the other from Northern Portugal (Gusmão et al. 1997).
In this study, new population data of this STR are reported for 17 populations of 3 major ethnic groups, Europeans, Asiatics and Africans. The data permitted documentation of some of the effects caused by the human spread through the world on the diversity patterns of this STR, and also to statistically explore the main mutational mechanism from which new alleles originate and to test the potential of this marker in phylogenetic studies.
Materials and methods
Data were independently obtained by two groups, one from the Section of Human Genetics, Institute for Health Research (INISA), University of Costa Rica, Costa Rica, and the other from the Institute of Biochemistry, Justus-Liebig University at Giessen, Germany. We decided to pool our results to achieve a wider perspective on the distribution and evolution of the polymorphism; therefore, some methodological differences in laboratory methods are to be noted.
Populations
Samples analyzed in Costa Rica are from 10 Amerindian populations, 1 group of Afro-Costa Ricans and 1 sample of the general Costa Rican population. Groups screened in Germany are 2 from Africa, 1 from Germany, 1 from Turks living or born in Germany and 1 from The Philippines. The populations were grouped into 3 major ethnic categories: (1) Asiatics, the Amerindians and the Filipinos; (2) sub-Saharan Africans (in this paper refered as Africans), the Namibian, Ugandan and the Afro-Costa Ricans; (3) Europeans, the Germans and the Turks. Data for the same marker from Northern Portugal reported by Gusmão et al. (1997) were added to the European group. The general Costa Rican population was considered as hybrid because of its multiethnic origin (Morera et al. 2001, 2003).
1. Asiatics
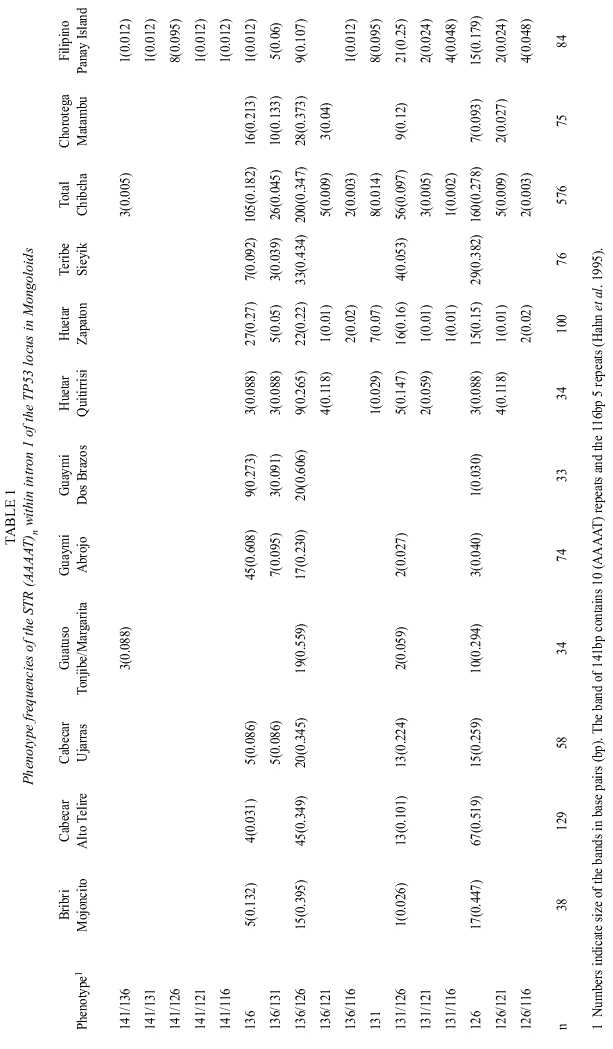
1.a. Amerindians: A total of 576 individuals belonging to six Chibchan-speaking tribal groups of Lower Central America (38 Bribri from Mojoncito, 129 Cabecar from Alto Telire, 58 Cabecar from Ujarras, 34 Guatuso from Tonjibe/Margarita, 74 Guaymi from Abrojo, 33 Guaymi from Dos Brazos, 34 Huetar from Quitirrisi, 100 Huetar from Zapaton, all in Costa Rica, and 76 Teribe from Sieyik, in Panama) were sampled between 1986 and 1998. The Chibchan Stock comprises 21 Central and South American aboriginal languages distributed along the Atlantic coasts of Honduras and Nicaragua, most of Costa Rica, Panama, almost the whole Colombian territory and the North of Venezuela (Constenla-Umaña 1991). An additional sample of 75 Amerindians was obtained in Matambu, Costa Rica, from a group whose ancestors belonged to an area of Mesoamerican (Central Amerind) cultural tradition and their now extinct language belonged to the Oto-Manguean Family (e.g. Newson 1987). Sampling methods were as described elsewhere (Barrantes et al. 1990).
1.b. Filipinos: 84 Filipino samples were obtained at the Panay Island, The Philippines, between 1992 and 1995. All are unrelated donors, collaborating as controls in a study on X-linked dystonia parkinsonism. Samples were cordially provided by Prof. Ulrich Müller, Institute of Human Genetics, Justus-Liebig University, Giessen.
2. Africans
2.a. Namibian: 99 samples from an Ovambo group, a Bantu tribe, were obtained in Northern Namibia, from voluntary blood donors to the Namibian Red Cross in 1994. Prof. Carsten Wolff, Halberstadt, kindly made them available to us.
2.b. Ugandan: 81 samples from a mixed population of Bantu people (Toro, Kiga and Tutsi), were obtained during a study on onchocercosis at the Kigoyera parish, Kabarole district, Western Uganda in October 1991. They were kindly contributed by Prof. Dietrich W. Büttner, Bernhard-Nocht Institute for Tropical Medicine, Hamburg.
2.c. Afro-Costa Rican: 193 samples of black donors were obtained at the seaport city Limón between 1999 and 2000. The origins of this group in Costa Rica dates back to 1872 when they were brought from Jamaica to labor in the construction of the railroad linking Limon with San Jose, the capital city of the country (Meléndez and Duncan 1979). Their exact African origins cannot be determined, nonetheless, it is certainly true that they are a mixture of peoples from different places (Meléndez and Duncan 1979, Lobo-Wiehoff and Meléndez-Obando 1997).
3. Europeans
3.a. Germans: The total sample adds up to 263 individuals: Bad Oeynhausen, North Rhine-Westphalia, 147 blood donors collected in 1998 and kindly provided by Prof. Carsten Wolff, Halberstadt; Hanover, 20 patients (bladder cancer) of the School of Medicine, sampled between 1995 and 1998, and, 96 donors from the whole area of Hanover, Lower Saxony, sampled between 1990 and 1992.
3.b. Turks: The sample, 88 individuals, was obtained in the Rhine-Main region, Germany, between 1994 and 1998. It comprises non-related patients afflicted with cystic fibrosis or persons suspected to be at risk of the disease. They were either immigrants from different regions of Turkey or individuals born to Turkish parents in Germany. The samples were kindly provided by Daniela Steinberger, Institute of Human Genetics, Justus-Liebig University, Giessen.
3.c. Portuguese: Data from Northern Portugal were taken from a published work (Gusmão et al. 1997) and added to the Europeans for the statistical analyses.
4. Hybrid
4.a. Costa Rican: 407 samples were obtained in 7 different regions of the country between 1996 and 1998.
Molecular screening
DNA extraction
DNAs analyzed in Costa Rica were extracted from whole peripheral blood (ACD) either with phenol-chloroform (Hermann and Frischauf 1987) or with NaCl (Miller et al. 1988). DNAs screened in Germany were also extracted from whole blood (EDTA) with phenol- chloroform.
PCR conditions
a. Conditions employed in Costa Rica. The oligonucleotide primers used were those described by Hahn et al. (1993). PCR-conditions were those of Ridanpää et al. (1995) but scaling down reaction volumes to 25µl instead of 50 µl.
b. Conditions employed in Germany: Two combinations of primers were used after Hahn et al. (1993, 2001). PCR conditions were according to Hahn et al. (2001).
Separation of PCR products
a. Conditions employed in Costa Rica. Native PAGE (3% stacking, 2cm below the comb level, and 10% resolving). Gels were stained with silver nitrate.
b. Conditions employed in Germany. Native and denaturing PAGE, and, denaturing capillary electrophoreses (Hahn et al. 2001).
Statistical analyses
Allele frequencies were estimated by gene counting and their standard deviations were calculated assuming a multinomial distribution; gene diversities were calculated after Nei (1987). Goodness of fit to the Hardy-Weinberg expectations, evaluated by a modified Fishers exact test (Guo and Thompson 1992), and fixation indexes, RIS , RST and RIT , and genetic distances were estimated according to Slatkin (1995) using the ARLEQUIN program (Schneider et al. 1997). A multidimensional analysis (Borg 1997) was performed using the STATISTICA for Windows, release 4.5 Package (StatSoft Inc. 1993).
The mutational origin of the allele-frequency distributions observed was assumed to follow a step-wise mutation model (Ohta and Kimura 1973, Moran 1975, Wehrhan 1975). The null hypothesis, that mutation occurs at one step events, was tested using the Markov-chains recursive method (Griffiths and Tavare 1994, Nielsen 1997) which takes the form of a likeli-hood ratio; i.e. the probability of a one-step model relative to that of a multi-step model (-2log[ L(
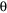
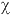
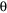
Results
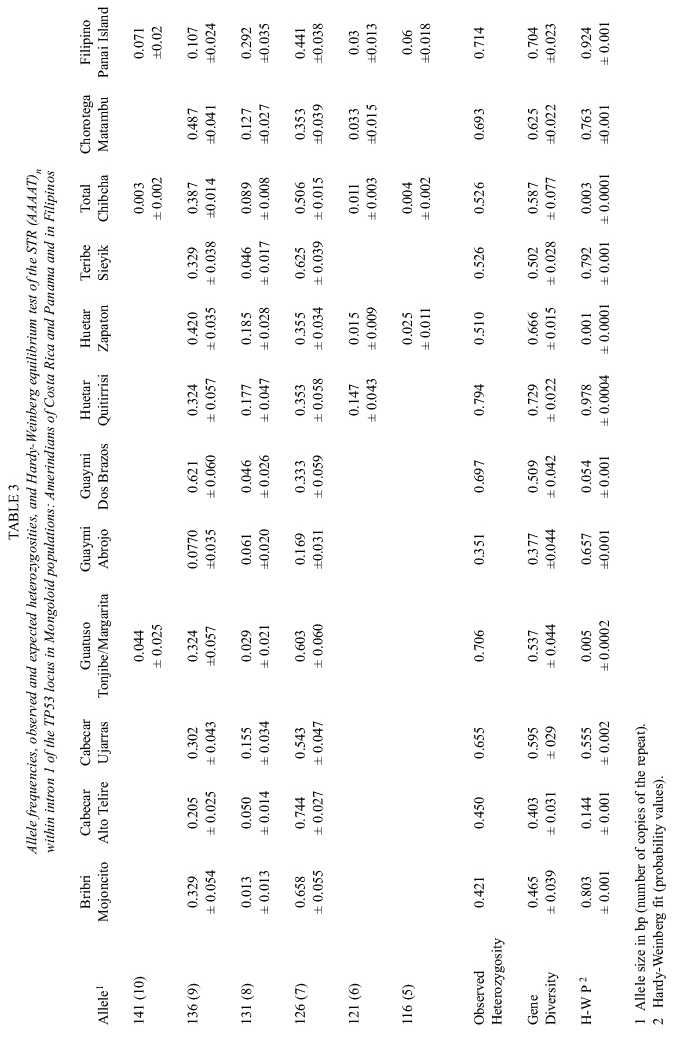
The absolute and relative phenotype frequencies obtained in the Asiatic group are presented in Table 1. Those found in the Africans, Europeans and the Hybrid population of Costa Rica are shown in Table 2. Their corresponding allele frequencies, observed and expected (gene diversities) heterozygosities and their Hardy-Weinberg fit are summarized in Tables 3 and 4.
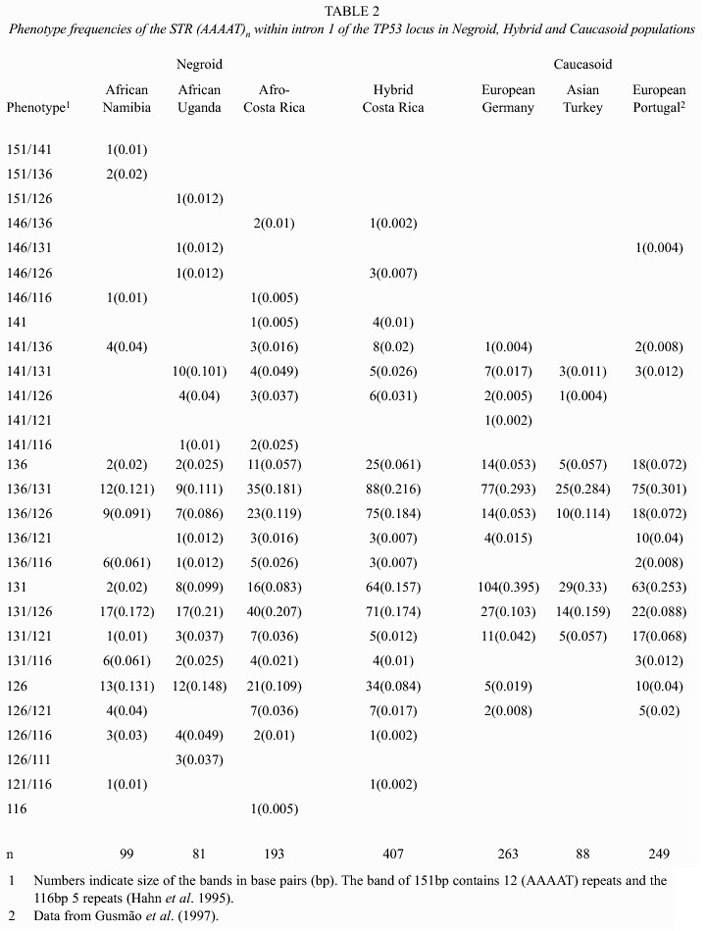
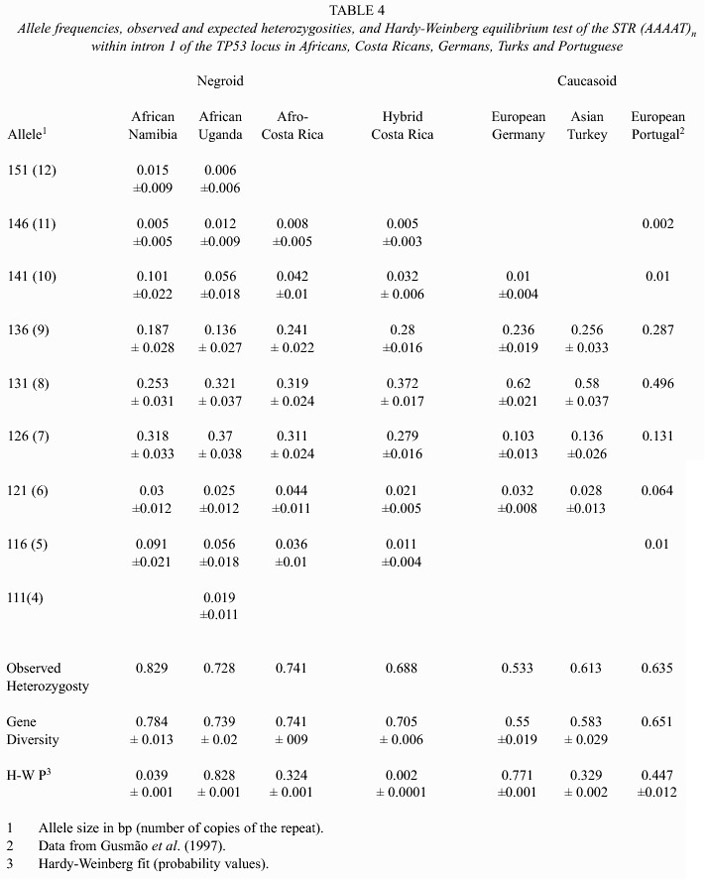
Although three alleles (126, 131, 136 bp) are the most frequent in all populations, the number of phenotypes and gene diversities follow patterns concordant with the geographic origin of each population as if they had spread from an African centroid. Africans, the oldest of them under this perspective, have, as would be expected, more alleles (7-9) more phenotypes (18-19) and higher gene diversities (0.739-0.784) than the other groups. Europeans, have 4-7 alleles, 6-14 phenotypes and gene diversities ranging from 0.55-0.705. Amerindians have 3-6 alleles, the number of phenotypes ranged between 3 and 12 and gene diversities between 0.377 and 0.729. However, setting apart the Guatuso, Huetar and Chorotega, tribes with the highest degree of interethnic admixture (Barrantes 1993), the rest of the tribes show an even lower diversity: 3 alleles, the number of phenotypes is 4 or 5 and the gene diversities range between 0.351 and 0.697. Filipinos have 9 alleles, 16 phenotypes and a gene diversity of 0.704, values intermediate between Africans and Europeans. The hybrid population of Costa Rica reflects, through its high diversity, 7 alleles, 20 phenotypes and a gene diversity of 0.705, its multiethnic origin.
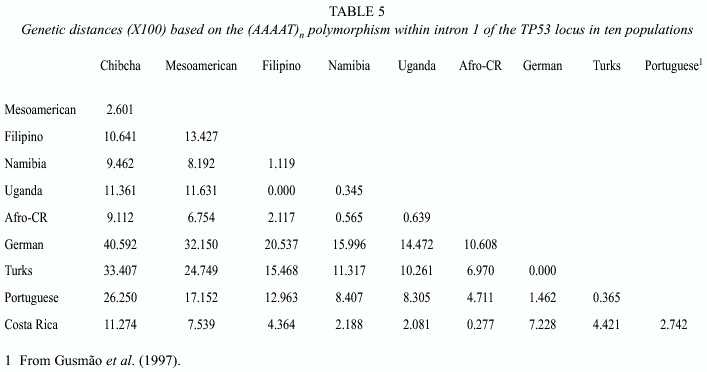
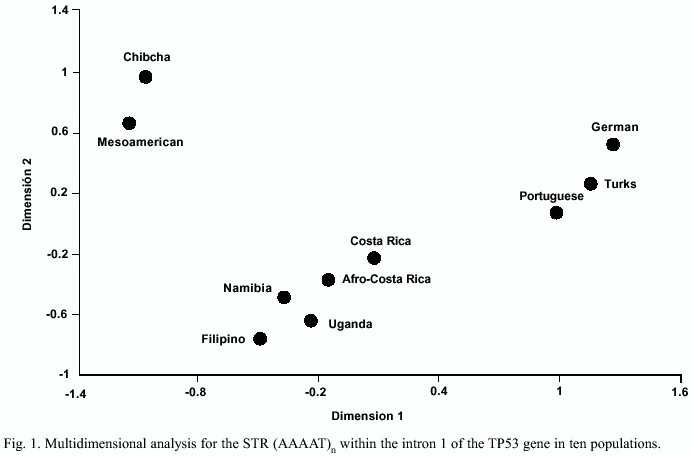
Genetic distances (Table 5) and a multidimensional analysis (Fig. 1) were performed to obtain a complementary perspective of the path followed by this STR during the divergence of these populations and to test the confidence of the data obtained from the samples analyzed (however, these statistics were not intended to estimate the actual genetic distances among the populations compared, i.e. they must be regarded in their qualitative sense). Populations clustered in three groups, an array in agreement with expectations according to their geographic origin and history: the Europeans, the Amerindians and a third one including the Africans, the Filipino and the hybrid population of Costa Rica.
The fixation indexes for the major groups, Europeans, Asiatics, Africans and hybrid were RIS = 0.0112 (p < 0.001), RIT = 0.0197 (p < 0.001) and RST = 0.0085 (p > 0.05). Thus variation is significant among individuals within subpopulations (RIS) as well as among individuals within the total of all populations (RIT ). In contrast, RST did not disclose any significant effect of population subdivision on the divergence of this STR at this level of population subdivision. The same was the case within Africans (RST = 0.0015, p > 0.05) and within Europeans (RST = -0.0005, p > 0.05). However, within the Asiatics R ST showed that subdivision in the three possible combinations has been significant: within the Chibchan (RST = 0.1411, p < 0.001), Chibchan total vs. Mesoamerican (RST = 0.0199, p < 0.05), and Chibchan total vs. Mesoamerican vs. Filipino (RST = 0.0324, p < 0.001).
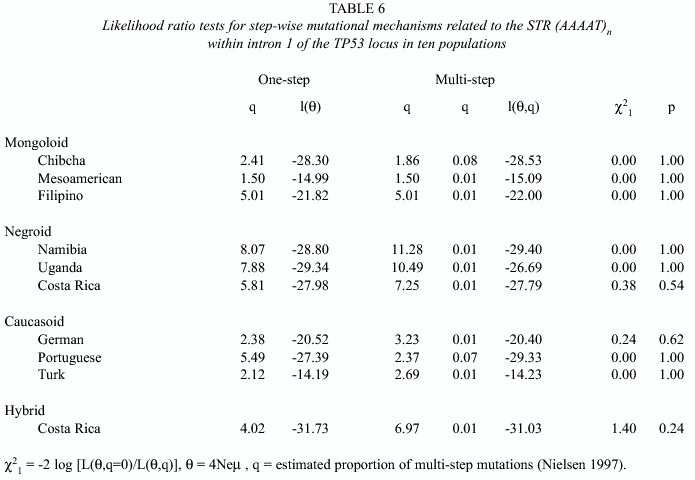
The data obtained also permitted a statistical exploration of the possible molecular mechanisms originating new alleles at this STR. Expansion or contraction in the number of tandem repeats could occur by single- or by multi-step events, i.e. the original allele could win or lose a single repeat or multiple repeats per mutational event. Table 6 summarizes like-lihood ratios for a one-step mutation model relative to a multi-step mutation model in ten populations (the Chibchan tribes were pooled as a single group). All ratios indicate that the single-step model better explains the observed allele-frequency distributions. Therefore, as q measures the joint effect of mutation, through its rate, and population structure, through Ne, it would be safe to postulate that mutation on this marker acts mainly by single steps.
Discussion
The characterization of genetic variation at an STR, the (AAAAT)n within intron 1 of the TP53 locus in 17 populations of 3 major ethnic groups, permitted the documentation of some of the effects of the spread of the human species through the world on its patterns of diversity. The subdivisions of ancestral populations, mainly by fissions, were events often associated with bottlenecks. Since the derived groups were small groups of related individuals, one of the main consequences during differentiation was the loss of alleles by genetic drift (Fix 1999). In the groups screened here the reduction of diversity follows an almost linear fashion from the African Continent: Africans, the oldest and most diverse, Europeans and Filipinos intermediate between the former and the youngest and less diverse Amerindians. Assuming a dispersal of the modern humans from an African centroid, this is an indication of the relative good representation of the samples screened and the discriminative power of the marker. The multidimensional analysis performed supports this interpretation, which is in very good agreement with worldwide studies based on many more genetic markers (e.g. Cavalli-Sforza et al. 1994). However, it must be kept in mind that it is based on a single marker, and, second, as noted by Harpending and Rogers (2000), measures of genetic distances of populations covering large geographic areas are often misleading.
Among the fixation indexes, RST , like genetic distances, also showed to be insensitive at major scales. A possible cause for this is the fact that three alleles (136/9, 131/8 and 126/7) are the most common in all populations and even though the size of the alleles ranged between 111/4 and 151/12, alleles other than the commonest rarely reached 10% or a higher frequency, which in turn could be the result of random fixation of alleles that can act differently on different loci. Some of these loci would have fixed different alleles in different parts of the world, as has been documented (Zerylnick et al. 1995, Deka et al. 1996, 1999). It could also be the case that they preserve the same alleles in all populations as has been shown for the CSF1R locus (Deka et al. 1999). Balancing selection cannot be ruled out given the important role of TP53 in genomic stability (Janus et al. 1999), and although the marker screened is located at an intron, increased instability of mRNA from alleles with motif-repeat numbers outside the range observed could affect the normal function of the gene. In addition, some alleles might have been indirectly selected by hitchhiking if they were in cis with alleles conferring increased fitness. The latter hypothesis could be tested extending the screening to haplotypic variation.
The Filipino and the hybrid Costa Rican population deserve comments aside. The Filipino shows an intermediate diversity between the Africans and Europeans and was unexpectedly placed at the multidimensional analysis closer to the Africans rather than to the Asiatics. Cavalli-Sforza et al. (1994) found a high diversity in this group using classical markers (blood groups and blood proteins) and explained the fact as a sign of a putative multiple origin (Negrito, Malayan and Chinese). In addition, the populations of Southeast Asia stem from an early split from the group that led to the other Asiatic groups (Cavalli-Sforza et al. 1994). Both facts, fusion of different ethnic groups each originating from a time when the ancestral (African) gene diversity was not diminished by bottlenecks could explain their higher diversity and closer proximity to the Africans than to the Amerindians. Regarding the high diversity value shown by the hybrid population of Costa Rica, it also reflects its multiethnic origin. Morera et al. (2003) estimated that the Costa Rican population has 61% European -mainly Spanish-, 30% Amerindian and 9% Africans genetic ancestry. Its nearer position to the African cluster is in accordance with that estimate. In the end, admixture can be seen as a regression to the ancestral conditions.
The data obtained also permitted a statistical analysis of the main molecular mechanism producing new alleles at this marker. The test favors a model of expansion or contraction in the number of tandem repeats by single-steps, although this does not rule out multi-step mutations. This conclusion is concordant with empirical data of Brinkmann et al. (1998) who analyzed 10 844 parent/child allelic transfers at 23 STRs and found that 22 out of 23 mismatches detected were single-step and only 1 a two-step.
Aknowledgments
We thank Omar Achí for his fine laboratory work. We also thank Carsten Wolff (Halberstadt, Germany), Ulrich Müller, Daniela Steinberger (Giessen, Germany) and Dietrich Büttner (Hamburg, Germany) for providing us the samples from Uganda, Namibia, The Philippines, Germany and Turkey. Support of Alfred Pingoud (Giessen) and Peter Lichter (Heidelberg), is also greatly acknowledged. This research was partially funded by the Universidad de Costa Rica (Grants 742-93- 903, 111-97-522 and 742-A1-502) and by the Consejo Nacional de Investigaciones Científicas y Tecnológicas de Costa Rica (CONICIT).
Resumen
Se estudió el polimorfismo del microsatélite (AAAAT)n del intrón 1 del gene TP53 en 17 poblaciones de 3 grupos étnicos: europeos, asiáticos, y africanos subsaharianos, así como de la población híbrida de Costa Rica (en total 1968 muestras). Tres alelos, 126/7 (pares de bases/ copias de la repetición), 131/8 y 136/9 fueron los más frecuentes en todas las poblaciones, aunque se observaron otros alelos usualmente a frecuencias menores al 10%. Las heterocigosis observadas variaron de 0.351 a 0.829. La distribución de la diversidad parece concordar con el origen geográfico de las muestras y con la historia de las poblaciones estudiadas. Una prueba estadística indica que el evento mutacional que más alelos nuevos produce en este marcador es el de un solo paso (expansión o contracción de una sola copia de la repetición). El índice de fijación RST mostró los efectos de la subdivision de poblaciones sólo dentro del grupo de los asiáticos y mostró falta de sensibilidad cuando los grupos comparados eran de niveles superiores de clasificación (europeos, asiáticos, y africanos) o cuando la comparación se hizo entre los grupos más antiguos (africanos y europeos).
References
Barrantes, R. 1993. Diversidad genética y mezcla racial en los amerindios de Costa Rica y Panamá. Rev. Biol. Trop. 41: 379-384. [ Links ]
Barrantes, R., P.E. Smouse, H.W. Mohrenweiser, H. Gershowitz., J. Azofeifa, T.D.Arias & J.V. Neel. 1990. Microevolution in Lower Central America: genetic characterization of the Chibcha-speaking groups of Costa Rica and Panama, and a consensus taxonomy based on genetic and linguistic affinity. Am. J. Hum. Genet. 46: 63-84. [ Links ]
Borg, I. 1997. Modern Multidimensional Scaling. Theory and applications. New York: Springer. [ Links ]
Brinkmann, B., M. Klintschar, F. Neuhuber, J. Hühne & R. Burkhard. 1998. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 62: 1408-1415. [ Links ]
Cavalli-Sforza, L.L., P. Menozzi & A. Piazza. 1994. The History and Geography of Human Genes. Princeton University, New Jersey. [ Links ]
Constenla-Umaña, A. 1991. Las lenguas del Área Intermedia. Introducción a su estudio areal. Editorial Universidad de Costa Rica, San José, Costa Rica. [ Links ]
Deka, R, P.P. Majumder, M.D. Shriver, D.N. Stivers, Y. Zhong, L.M. Yu, R. Barrantes, S.J. Yin, T. Miki, J. Hundrieser, C.H. Bunker, S.T. McGarvey, S. Sakallah, R.E. Ferrell & R. Chakraborty. 1996. Distribution and evolution of CTG repeats at the myotonin protein kinase gene in human populations. Genome Res. 6: 142-154. [ Links ]
Deka, R, M.D. Shriver, L.M. Yu, E. Mueller-Heidreich, L. Jin, Y. Zhong, S.T. McGarvey, S.S. Argawal, C.H. Bunker, T. Miki, J. Hundrieser, S.J. Yin, S. Raskin, R. Barrantes, R.E. Ferrel & R. Chakraborty. 1999. Genetic variation at twentythree microsatellite loci in sixteen human populations. J. Genet. 78: 99-121. [ Links ]
Fix, A.G. 1999. Migration and Colonization in Human Microevolution. Cambridge University, Cambridge. [ Links ] Futreal, P.A., J.C. Barret & R.W. Wiseman. 1991. An Alu polymorphism intragenic to the TP53 gene. Nucl. Acids Res. 19: 6977. [ Links ]
Gaspar, P.A., M.H. Hutz, F.M. Salzano & T.A. Weimer. 2001. TP53 polymorphisms and haplotypes in South Amerindians and neo-Brazilians. Ann. Hum. Biol. 28: 184-194. [ Links ]
Gaspar, P.A., M.H. Hutz, F.M. Salzano, K. Hill, A.M. Hurtado, ML. Petz-Erler, L.T. Tsuneto & T.A. Weimer. 2002. Polymorphisms of CYP1A1, CYP2E1, GSTM1, GSTT1, and TP53 genes in Amerindians. Am. J. Phys. Anthrop. 119: 249-256. [ Links ]
Griffiths, R.C. & S. Tavare. 1994. Simulating probability distributions. Theor. Pop. Biol. 46: 131-159. [ Links ]
Guo, S. & E. Thompson. 1992. Performing the exact test of the Hardy-Weinberg proportion for multiple alleles. Biometrics 48: 361-372. [ Links ]
Gusmão, L., M. J. Prata, A. Amorim, F. Silva & I. Bessa. 1997. Characterization of four short tandem repeat loci (THO1, VWA31/A, CD4, and TP53) in Northern Portugal. Hum. Biol. 69: 31-40. [ Links ]
Hahn, M., J. Serth, R. Fislage, H. Wolfes, R. Allhoff, U. Jonas & A. Pingoud. 1993. Polymerase chain reaction detection of a highly polymorphic VNTR segment in intron 1 of the human p53 gene. Clin. Chem. 39: 549-550. [ Links ]
Hahn, M., R. Fislage & A. Pingoud. 1995. Polymorphism of the pentanucleotide repeat d(AAAAT) within intron 1 of the human tumor suppressor gene p53 (17p13.1). Hum. Genet. 95: 471-472. [ Links ]
Hahn, M., J. Wilhelm & A. Pingoud. 2001. Influence of fluorophor dye labels on the migration behavior of polymerase chain reaction-amplified short tandem repeats during denaturing capillary electrophoresis. Electrophoresis 22: 2691-2700. [ Links ]
Harpending, H. & A. Rogers. 2000. Genetic perspectives on human origins and differentiation. Ann. Rev. Genomics Hum. Genet. 1: 361-385. [ Links ]
Hermann, B.G. & A.M. Frischauf. 1987. Isolation of genomic DNA. Methods Enzymol. 152: 180-183. [ Links ]
Hesketh, R. 1997. The Oncogene and Tumor Suppressor Gene Facts Book. Academic, London. [ Links ]
Isobe, M., B.S. Emanuel, D. Givol, M. Oren & C.M. Croce. 1986. Localization of the human p53 tumor antigen to band 17p13. Nature 320: 84-85. [ Links ]
Janus, F., N. Albrechtsen, I. Dornreiter, L. Wiesmüller, F. Grosse & W. Deppert. 1999. The dual role model for p53 in maintaining genomic integrity. Cell. Mol. Life Sci. 55: 12-27. [ Links ]
Lobo-Wiehoff, T. & M. Meléndez-Obando. 1997. Negros y blancos. Todo Mezclado. Editorial de la Universidad de Costa Rica, San José, Costa Rica. [ Links ]
Meléndez, C. & Q. Duncan. 1979. El Negro en Costa Rica. Editorial Costa Rica, San José, Costa Rica. [ Links ]
Miller, S.A., D.D. Dykes & H.F. Polesky. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl. Acids Res. 16: 1215. [ Links ]
Moran, P.A. 1975. Wandering distributions and the electrophoretic profile. Theor. Pop. Biol. 8: 318-330. [ Links ]
Morera, B., R. Marín-Rojas & R. Barrantes. 2001. Análisis de varios marcadores genéticos clásicos en la población de Costa Rica. Rev. Biol. Trop. 49: 1237- 1252. [ Links ]
Morera, B., R. Barrantes & R. Marin-Rojas. 2003. Gene admixture in the Costa Rican population. Ann. Hum. Genet. 67: 71-80. [ Links ]
Nei, M. 1987. Molecular Evolutionary Genetics. Columbia University, New York. [ Links ]
Newson, L. 1987. Indian Survival in Colonial Nicaragua. Norman, University of Oklahoma. [ Links ]
Nielsen, R. 1997. A likelihood approach to population samples of microsatellite alleles. Genetics 146: 711-716. [ Links ]
Ohta, T. & M. Kimura. 1973. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 22: 201-204. [ Links ]
Ridanpää, M., S. Anttila & K. Husgafvel-Pursiainen. 1995. Detection of loss of heterozygosity in the p53 tumor suppressor gene using a PCR-based assay. Path. Res. Pract. 191: 399-402. [ Links ]
Schneider, S., J.M. Kueffer, D. Roesli & L. Excoffier. 1997. Arlequin version 1.1. A software for population genetic data analysis. Genetic and Biometry Laboratory, Department of Anthropology. University of Geneva, Switzerland. (Available at: http://lgb.unige.ch/arlequin/). [ Links ]
Själander, A., R. Birgander, A. Kivelä & G. Beckman. 1995. p53 Polymorphisms and haplotypes in different ethnic groups. Hum. Hered. 45: 144-149. [ Links ]
Själander, A., R. Birgander, N. Saha, L. Beckman & G. Beckman. 1996. p53 polymorphisms and haplotypes show distinct differences between major ethnic groups. Hum. Hered. 46: 41-48. [ Links ]
Slatkin, M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457-462. [ Links ]
Wehrhan, C.F. 1975. The evolution of selectively similar electrophoretically detectable alleles in finite natural populations. Genetics 80: 375-394. [ Links ]
Zerylnick, C., A. Torroni, S.L. Sherman & S.T. Warren. 1995. Normal variation at the myotonic dystrophy locus in global human populations. Am. J. Hum. Genet. 56: 123-130. [ Links ]
Internet references
GDB (TM). 2003. Human Genome Database [database online]. Available from Internet: URL http://www.gdb.org (Downloaded: 11 Aug. 2003; GDB node: The Netherlands: http://gdb.cmbi.kun.nl/gdb/ ) GDB Data Type: Gene; object name: TP53; Accession ID: GDB: 120445. [ Links ]
OMIM (TM). 2000. Online Mendelian Inheritance in Man. Johns Hopkins University, Baltimore, MD, MIM 191170. Date of last modification 7/24/2003. (Downloaded: http://www.ncbi.nlm.nih.gov/htbinpost/ Omim/dispmim?191170#.). [ Links ]