Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.50 n.2 San José Jun. 2002
Edgardo Moreno
Programa de Investigación en Enfermedades Tropicales (PIET), Escuela de Medicina Veterinaria, Universidad Nacional, Heredia, Costa Rica. Fax: (506) 2381298, E-mail: emoreno@irazu.una.ac.cr
(ReceivedI4-IV-I996. Corrected 9-X-I996. Accepted30-X-1996.)
Abstract: The bacterial species concept was examined within the framework of plant and animal associated α-2 proteobacteria, taking into consideration the phylogenetic, taxonomic and biological approaches as well as the microbiologists perception. The virtue of the phylogenetic approach is that it gives an evolutionary perspective of the bacterial lineage; however the methods used possess low resolution for defining species located at the terminal branches of the phylogenetic trees. The merit of the taxonomic approach is that species are defined on the basis of multiple characteristics allowing high resolution at the terminal branches of dendograms; its disadvantage is the inaccuracy in the earlier nodes. On an individual level, the qualitative biological characteristics used for the definition of species frequently reveal shortcomings because many of these properties are the result of coevolution, parallel evolution or the horizontal transfer of genes. Nevertheless, when considered together with !be phylogenetic and taxonomic approaches, important uncertainties are discovered: these must be weighed if a practical definition of bacterial species is conceived. The microbiologists' perception is !be criterion expressed by a group of sponsors who, based on scientific and practical grounds, propose a new bacterial species. The success of this new proposal is measured by its widespread acceptance and its permanence. A difficult problem concerned with defining bacterial species is how to distinguish if they are independent evolutionary units or if they are reticulate evolutionary units. In the first case the inherence is vertically transmitted as a result of binary fission and clonal expansion. This may be !be case of some animal cell associated bacteria in which recombination appears to be precluded or exceptional. In the second case adaptive changes occurring within an individual can be horizontaIly transferred to many or all group members. This seems to be the condition of many intestinal and plant associated bacteria. Genetic drift and speciation in clonal bacteria will depend almost exclusively on mutation and internal genetic rearrangement processes, whereas speciation in reticulate bacteria will depend not only on these processes but in their genetic interactions with other bacterial strains. This uncertainty, which corresponds to the evolutionary process, is at the same time one of the key factors in defining a bacterial species.
Key words: Bacterial species, Proteobacteria, review, Brucella, Bartonella, Ochrobactrum, Afipia, Agrobacterium, Rhizobium, Phyllobacterium, Rhodopseudomonas, Bradyrhizobium.
"They, and every beast after his kind, and all the cattle after their kind, and every creeping thing that creepeth upon the earth after his kind, and every fowl after his kind, every bird of every sort. GENESIS 7: 14 And they went in unto Noah into the ark, two and two of all flesh, wherein is the breath of life. GENESIS 7: 15 And they that went in, went in male and female of all flesh, as God had commanded him: "and the Lord shut him in. GENESIS 7: 16".
The job assigned to the mythical Noah was monumental, not only because he had to calculate how to accommodate all species of animals in a finite ark size (300 cubits long x 50 cubits wide x 30 cubits high) but also due to the enormous and exhausting duty of sorting and identifying in a brief death line (two months and seventeen days) the different species of animals, their sexes, their feeding habits and above all, to estimate their chances for survival. Conveniently for Noah, his work in systematics concluded after the ark's door was closed. However, Noah's task is far from complete and the question concerning the correct identification of species and where to locate them is still a major puzzle.
The proximity of an ecological cataclysm, where human beings actively participate in the extinction of a considerable number of species, has alerted many instances in the relevance of sampling and classification of living creatures. Due to the fact that plants and animals are more conspicuous and in consequence more unveiled to our prejudices regarding beauty, ugliness and usefulness, most of the new "Noah's arks" have been devoted to the census of these biodiversities with little or marginal attention to the microbial world. If we consider that the primordial living systems (and very probably the ultimate) were assembled almost exclusively by bacteria or bacteria-like organisms, and that practically all the genetics as well as the synthetic, catabolic and regulatory biochemical reactions were developed by these organisms and are still concentrated in bacteria, then it is obvious that we must give to this world its place in our "modem arks". Furthermore, by considering the simple but relevant fact that practicalIy all the eukaryotic systems studied possess or interact with at least one unique eubacteria (including endosymbionts), it is reasonable to propose that the bacterial world is not only the most abundant but the most diverse of all.
Shortcomings on the isolation and culture of a great number of bacteria (mainly the intracellular and the "extremist") have restricted the correct quantification and characterisation of these organisms. Even though this problems have been partially solved by the advent of recent molecular techniques devoted to the amplification and sequencing of genes from bacteria which are difficult to isolate or to cultivate under controlled conditions, the truth is that classification of bacteria into species is .a constant drawback (Krieg, and Holt. 1984). Today it is clear (at least for some of us) that the success of all conservation programs, biodiversity projects, bacterial collections, microbial ecologists, medical microbiologists, plant pathologists, food microbiologists, epidemiologists and geneticists depends on the correct identification of those extant units known as bacterial species for which protection, compilation, conservation, control, destruction or selection is being sought.
Since the publication of Darwin's masterpiece "On the Origin of Species" in 1859, the definition of species concerning animals and plants has been extensively debated by a great number of scientists (O'Hara 1993 and 1994, Avise 1994,). In contrast, the bacterial world was not subjected to the same detailed analysis for more than one century. This unfortunate fact was mainly due to the absence of data sustaining the evolutionary history of bacteria (Woese 1987). The advent, during the last two decades, of concise phylogenetic and taxonomic studies based on molecular sequences and structures has definitively contributed to incorporating the bacterial world within the constraints of evolutionary studies (Woese 1994). As with other organisms, the distinctiveness of bacteria requires an idiosyncratic analysis for the concept of species. However, we must distinguish between the various bacterial species concepts and the different character sets used to define species in that the former refers to a philosophical position and the latter refers to the characteristics used by the classification methods. Furthermore, the different systems of classification force us to a particular model for representing the dispersion of species which in most cases overlap with the different definitions (O' Hara 1993). In order to obtain a correct perspective on this, I will first of all discuss the bacterial species concept within the framework of the different species views which at the same time are related to the different character sets used to distinguish the various species. Up to the present, three different views on that which constitutes a species and two perceptions dominate our present knowledge: the phylogenetic species concept championed by Cracraft (1983), the taxonomic species concept (Staley and Krieg 1984), the biological species concept (Dobzhansky 1937), the microbiologists' perception (Pitt 1994, Sneath 1984b) and the fixed species perception reviewed by Schadewald (1986). The three concepts have strengths and weaknesses, each emphasises different aspects of the evolutionary process and are within the boundaries of science. Because the taxonomic and phylogenetic concepts are based on numerical analysis (Sneath 1984a, Woese 1994, Williams et al. 1995), they are the most distinctly understood. The biological species concept may be considered the ultimate definition, but due to the complexity involved in understanding the biology of a bacteria regarded as qualitative characters, it has been relegated to last among the three scientific views (Krieg and Holt 1984). In this essay I attempt to rescue the value of the biological species concept in the framework of the bacterial population structure. Other definitions such as the concordance (Avise and BaIl 1990), recognition (Paterson 1985), cohesion (Templeton 1989) and evolutionary (Simpson 1951) species concepts, which have been used for the definition of animal and plant species, are either included within ¡he phylogenetic, taxonomic and biological concepts or are not relevant within the context of a practical bacterial species definition. The two perceptions are anthropocentric and non scientific in nature. Nevertheless, the microbiologists perception must be taken into account since sooner or later "a classification that is of little use to the microbiologist, no matter how fine a scheme or who devised it, will be ignored or significantly modified" (Staley and Krieg 1984). The fixed species perception may be the most popular view among laymen. Since this perception is not amenable to scientific treatment or open to discussion outside the limits of creationism (Schadewald 1986), I will not consider it in further discussions.
On attempting to define a concept of a bacterial species, different and sometimes contrasting views must be considered. However, the concepts should be limited to the constraints of what is believed to be a natural system of classification. In short, the bacterial species must be catalogued in a system commensurate with the currently accepted theory of evolution. This includes a concept that takes into account the important role of the reproductive isolation of organisms and the horizontal transfer of genes as different and complementary modes of bacterial speciation (Syvanen 1994). Understanding of this is expected to have a profound influence on the future of bacterial systematics.
The Phylogenetic Approach
A phylogenetic species may be understood as 'the smallest diagnosable cluster of individual organisms within which there is a parental pattern of ancestry and descendent" (Cracraft 1983). This concept has the advantage that species are defined as the terminal organism in a lineage which has an ancestor common to other terminal organisms, represented by the node joining of their two branches; thus avoiding the meaningless classical ranked classification into kingdoms, phyla, classes, and families. In the light of this definition, debates concerning bacterial ranks above the genus level are trivial and unimportant (O'Hara 1994). A common problem with this concept involves determining how monophyly is to be recognised and how to distinguish gene phylogenies from pedigrees (Avise 1994).
The phylogenetic species concept is commonly based on two different types of data generated by different techniques: the comparison of characters between fossils and the extant species and the comparison of molecular sequences of the different organisms. Since the availability of bacterial fossils is scarce (Ochman and Wilson 1987), the evolutionary history of these microorganisms has been mainly reconstructed on the basis of molecular sequences which are best represented in the form of evolutionary trees or bushes (Olsen et al. 1994). Questions, such as if a gene can represent an organism and which bacterialgenes best represent the evolutionary history of bacteria, have been a matter of dispute among evolutionists (Forterre et al. 1993). The weight of evidence generated during the last few years strongly suggests that some genes can in fact represent the phylogeny of bacteria (Woese 1987, Olsen and Woese 1993, Olsen et al. 1994). The search has been focused on genes that perform central functions, that is, "the entities most tightly interwoven into the fabric of the cell itself' (Olsen and Woese 1993). In this sense, bacteria phylogenies based on the sequences of conserved molecules such as ribosomal RNAs, chaperone proteins, cytochrome c, nitrogenase, elongation factor, ATPase synthase and glutamine synthetase, among others (Ochman and Wilson 1987, Kumada et al. 1993, Ludwing et al. 1993, Olsen et al. 1994, Wallington and Lund 1994, Woese 1994) have greatly contributed to discerning the evolutionary relationships between bacteria, and therefore have substantially helped to identify different bacterial groups. Unfortunately, the same properties that make these molecules suitable for phylogenetic classification at a certain level, make them poor tools for defining bacterial species within close sister clades. For instance, I agree with the concept that there are genes (such as those cited above) which can approach the phylogeny of an organism; however, I also understand that at the species level they do not have enough resolution for defining bacterial species as separate entities (Stackebrandt and Goebel 1994). Furthermore, some of these bacterial genes are not single units but rather are duplicated in the same chromosome (Dryden and Kaplan 1990, Viale ei al. 1994) or repeated in a second (different) chromosome of some bacteria (Allardet-Servent et al. 1988, Dryden and Kaplan 1990) and are therefore subjected to different positional constraints that may promote diversity within the same group. It can be argued that, given a gene (or the entire genome), one can always discern between close bacterial relatives and resolve them into species. The main problem with this is the absence of limits in the prediction of a terminal taxa: if one tries hard enough, sequence differences can be found to diagnose virtually any bacterial population. The "pendular movement machine" operated by the "lumpers" and the "splitters" of bacterial taxonomy depends on the data provided by them, generating from one to an ample number of species.
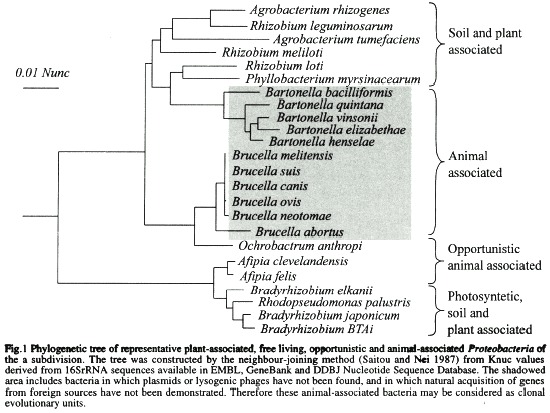
For the sake of our discussion, let us consider the phylogenetic tree in Fig. 1 constructed by the neighbor-joining method (Saitou and Nei 1987) from Knuc values derived from 16SrRNA sequences. All the terminal taxa represented in this tree correspond to phylogenetic close relatives of Gram negative bacteria assigned to the α-2 subdivision of Proteobacteria (Olsen et al. 1994). On consulting the topology of the tree we can at first glance resolve that in this case the traditional concept of Family established by ranked classification is a complete catastrophe in the sense that members of the Rhizobiaceae (Agrobacterium spp. Rhizobium spp. Phyllobacterium spp. and.Bradyrhizobium spp.) are not only intermingled with bacteria originally included within the Rickettsiaseae (Bartonella quintana and B. visonii, former Rochalimae species), Bartonellaceae (B. bacilliformis) and members of "unknown" affiliation (Brucella spp. and Afipia spp.), but are also themselves mixed up (e.g. R. meliloti is closer to A. tumefasciens than to other members of the same genus). Furthermore, Bradyrhizobium spp. belongs to a different branch of association, together with Rhodobacter sphaeroides, the latter bacteria being considered a member of the Rhodospirillaceae family (Imhoff and Trüper 1984). In this respect, classification based on sequence analysis has clearly contributed to discerning sister clades and appropriately defining groups among the α-2 proteobacteria, particularly at the genus level (Stackebrandt and GoebeI 1994). In addition, it has made us look more carefully at the common factors of bacteria belonging to the same lineage, sometimes in spite of the diverse biology displayed by sister organisms (Moreno 1992). However, at the species level the tree shown in Fig. 1 has poor resolution for some strains: this is clearly illustrated by the compact clades demonstrated in the Brucella, Bartonella and Bradyrhizobium terminal taxons, whose similarities are above 97%. As a result of this problem a combination of analysis of idiosyncratic sequences and numerical analysis of phenotypic characteristics has been undertaken by several researchers in order to discern between closely related táxones (Wayne et al. 1987, Ficht et al. 1996).
The Taxonomic Approach
The taxonomic species concept is a definition clearly linked to its method. For many years, the aim of the microbial taxonomist has been to devise a consistent set of techniques for the classification of bacteria into different ranks. The most common procedures are based on the analysis of phenotypic features, biochemical characteristics, genotypic similarities and immunochemical reactions. Modern bacterial evolutionary units taxonomists sort the available figures using numerical analysis and synthesise them in the form of clades or dendogrames (Gao et al. 1994, Williams et al. 1995). The basic taxonomic unit generated by this analysis is defined as a taxospecies (Sneath 1984a). The same procedures used to determine a species are employed to further characterise the terminal taxons into subspecies, biotypes, serotypes, chemotypes, phagotypes, phenotypes and so on. The virtue of the taxonomic proposition is that it is defined in a polyphasic manner allowing high resolution at the terminal branches of dendograms; its drawback is the inaccuracy in the earlier nodes of the branches (e.g. compare phylogenetic trees from Yanagi and Yamasato [1993] with phenic dendograms generated by Gao et al. [1994] ) This is mainly due to the fact that many characteristics displayed by the different bacteria (e.g. serological cross reactivity, staining properties, resistance to antibiotics, etc.) may be the product of coevolution, parallel evolution or the result of horizontal transfer of genes. However, the combination of phylogenetic grouping based on sequence comparisons with taxonomic classification is a very powerful method for approaching a practical definition of the bacterial species (Stackebrandt and GoebeI 1994). In spite of all this advantages, this proposed polyphasic approach possesses some drawbacks as will be demonstrated in the following paragraphs.
For instance, Fig. 1 shows that the plant endosymbiont B. japonicum is phylogenetically closely related to the Bradyrhizobium strain BTA1; however, the latter is photosynthetic while the former is not. Since this characteristic is chromosomal coded and unlikely to be horizontally transferred, these two bacteria could be tentatively classified as different species. Similarly the phylogenetically closely related Bartonella species can be differentiated into ten individual species according not only to different phenotypic, chemotypic and immunochemical criteria, but more importantly to DNA-DNA reassociation values (Birtles et al. 1995). The latter procedure has been pragmatically established as a "superior method" for the classification of species (Stackebrandt and Goebel 1994). Nevertheless, our experience on studying the microbiological world has taught us that reality challenges the imagination. Not without lengthy debate and a considerable input of energy (and some bile), the genus Brucella is believed to comprise six species (CorbeI1989, Meyer 1990), in spite of the fact that phylogenetic analysis based on conserved molecules such as 16SrRNA similarity and DNA relatedness values indicate figures above 99% and 98% respectively (Fig. 1 and Verger et al.[1985]). The reason is not only because the different Brucella species may be resolved on the basis of DNA restriction patterns (AlIardet-Servent et al. 1988, Fekete et al. 1992), phylogenetic analysis of idiosyncratic sequences (Ficht et al. 1996) as well as phenotypic, chemotypic and antigenic characteristics (Moreno 1992), but more important because of the distinct biological behaviour of the species that distinguishes one from the other (Corbel 1989, Meyer 1990). In this particular case the DNA-DNA reassociation technique has been considered an "inferior method" compared to the biological characteristics of these bacteria and to other assays (Meyer 1990).
The Biological Approach
In contrast to the numerical analyses used in the phylogenetic and taxonomic studies for the definition of bacterial species, the biological concept is based upon the identification of fundamental qualitative characteristics. In modern biological usage, species are defined as a particular group of organisms which retain their distinctness from other kinds in nature over a period of successive generations. When referring to sexually reproducing organisms, "distinctness" commonly designates the ability of members of the same species to interbreed freely, whereas interbreeding between members of different species is prevented or restricted by natural causes. In another context, "distinctness" means isolation in a non interacting different habitat. In spite of some shortcomings (Ayala 1976), the biological concept still has a profound impact on the recognition of animal and plant species (Avise 1994). One important difference between this consideration and the previous definitions is its predictive nature, where time projects into the future from the moment in which a particular species is recognised. Such a claim about the future may of course turn out to be false since the separation between two or more species may be temporary or permanent depending upon the way the world goes (Danto 1985). With respect to bacteria, the biological species concept has been defined on the basis of certain distinctive biological characteristics such as the basic comprehension of the life cycle (preferred habitat, potential habitat, preferred host, secondary hosts, vectors, reservoirs, interaction with other bacteria, symbiosis, commensalism, parasitism, saprophytic life and modes of transmission), bacterial physiology (metabolism, generation time, optimal growth temperature, defence mechanisms, offensive mechanisms, reparation systems and internal rearrangement of genes), and reproductive behaviour (vertical vrs. horizontal inherence, conjugation, transduction, retrotransposition, transformation and capsduction). The bacterial specie defined on the basis of qualitative biological characteristics is sometimes referred to as genospecies (Sneath 1984a). However, on finding that similar biological traits may occur due to convergent or parallel evolution or may be acquired by the horizontal transfer of genes (Lambert et al. 1990), the traditional biological species concept must be modified in favour of a more versatile notion.
Experience has demonstrated that biological characteristics themselves are of little use in defining bacterial species (Weisburg et al. 1989). However, when phylogeny and taxonomy are considered, the biological properties shown by a bacterium contribute to resolving important uncertainties. For instance, B. bacilliformis, the etiological agent of Oroya Fever, can be recognised from other Bartonella species (Fig. 1), not only by its clear taxonomic features (Brenner et al. 1993, Birtles et al. 1995) but because this bacterium, which is restricted to a small geographical area in Peru, is a human pathogen exclusively transmitted by one distinct species of sand flies (Gray et al. 1990). Since humans are the only vertebrate host for B. bacilliformis, and our species reached the highlands of Oroya no more than 12,000 years ago, it is reasonable to propose that this particular parasitic bacterium emerged in its present form around that time. Likewise, in spite of the indistinguishable DNA-DNA reassociation similarity index shown by the different Brucella species (Verger et al. 1985), they can be recognised by their biological attributes, such as host range and pathogenicity (Corbel 1989, Meyer 1990). Sheep are the only animal host from which B. ovis has been recovered, whereas the wood-rat Neotoma lepido. is the only known host for B. neotomae. Furthermore, B. neotomae is non pathogenic for its host; in contrast B. ovis primarily affects males (causing ram epididymitis) and has seldom been encountered among ewes. Dogs are the natural host for B. canis; this bacterium seldom infects other species, and when it does infection occurs without any epidemiological importance since the bacteria cannot be transmitted. Similarly, the host animals for B. abortus, B. melitensis and B. suis are bovines, caprines and suines respectively (with the only exception of B. suis biotype 4, which seems to be specific for reindeer, and therefore defined by some taxonomist as a distinct species). These three Brucella species are very aggressive pathogens which may infect other hosts; however, when this occurs infection is often terminal and of minor epidemiological importance. What makes the biological attributes of these microorganisms important in the context of the species definition is the fact that under natural conditions the distinct biochemical, phenotypic, immunochemical and genomic attributes that identify each of the different Brucella species have remained constant in space and time (Corbel 1989, Meyer 1990). These clear-cut examples force us to consider the qualitative biological characteristics as a valid alternative in the scheme of a species definition. Nevertheless, with respect to other bacteria we must be cautious ,in our judgement since distinct biological behaviour may depend on the presence of accessory genetic elements.
The Microbiologists' Perception
Carolus Linnaeus himself admitted that his classification did not always categorise the most similar things together (including humans and apes who were recorded as separate taxons), but he felt that his system was very well adapted to the interests of plant growers and botanists (as well as clergies) of his time. The virtue of the binomial system of nomenclature established by this Swedish naturalist in 1753 has been its clarity and simplicity. These attributes constitute the main reason why the linnaen system is still used for naming all the organisms within the three domains of living things: Bacteria, Archea and Eucarya (Woese 1987). Prior to Linnaeus' Species Plantarum publication, the names of the species were in reality short Latin descriptions, usually including several words. This complicated designation attempted to present the main characters by which the species could be recognised. The wisdom of Linnaeus was not only to create a comprehensible classification system, but more importantly, a useful one.
The development of a natural system, in which living things are classified according to the totality of their similarities and differences and evolutionary traits, has occupied the minds of post-linnaen investigators to this day, and it will very probably never be satisfactorily completed. The definition of a bacterial species furnishes the basis of our knowledge of it, but that same knowledge also helps us to use it more effectively. The art of the systematic bacteriologist lies not only in the correct use of methods and in the construction of elaborated theories to reach the "final" goal of successfully defining an entity that he or she thinks is a species, but also in convincing the (scientific) community that this definition is more appropriate to the reality of their disciplines. Although, some general rules for devising a valid classification may be established by many, eventually only a few will be responsible for actually naming a species. In this respect the microbiologist species perception is sustained upon the criteria expressed by a sponsor or a group of sponsors (experts, scientists, professionals, organisations) who based on scientific grounds (phylogenic, taxonomic and biological studies) and on practical parameters (epidemiology, production, potential use, risks, ranking orders, and tradition) propose a new bacterial species.
In considering the phylogenetic trees, taxonomic dendograms or biological separation based on what is believed to be the most natural system of classification for designation of species, the microbiologists must be aware that at higher systematic levels these representations are retrospective, but close to the species level they are prospective, that is, dependent upon the interpretation of the future (O'Hara 1993). This is commonly referred as the "problem of prospective narrative predication" (Danto 1985) since the separation between two bacterial populations as distinct species depend upon future contingencies which may be estimated but not predicted. As a consequence, it is logically impossible for all the species concepts to be applied with certainty to the present because they all depend upon the future(O'Hara 1993). However, taking into consideration the relatively short life span of humans, microbiologists are confronted with the dilemma of reaching a consensus for defining a bacterial species that is useful for all purposes. In this respect the data obtained by the various approaches must be weighed in the light of the different bacterial species concepts. The success of this enterprise will be measured by its widespread acceptance and its permanence. Therefore, the closest approximation to a bacterial species definition will be one that is widely accepted by scientists and practising professionals and which resists the inexorable vicissitudes of time. This problem is similar to the disadvantage of the relativistic theory when considering space and time in our daily lives with respect to the Newtonian approach which, in spite of its drawbacks, is more practical for ibis purpose.
From its initial "baptism" as Rickettsiae quintana, followed by its "re-baptism" as Rochalimae quintana, succeeded by its "superbaptism" as Bartonella quintana, this bacterium has changed none of its attributes (although new ones have been discovered), apart from its name (Brenner et al. 1993). The fast and widespread acceptance of the new nomenclature by the scientific community, veterinarians and physicians (Breitschwerdt et al. 1994, Kordick and Breitschwerdt 1995), in many ways reveals the success of the changes, because the new classification is in accordance with the attributes of the bacterium. As in a fairytale happy ending, phylogenetic taxonomic and biological studies have greatly contributed to joining this bacterum with its sister species (Fig. 1). However, other bacterial groups are not as fortunate and despite extensive studies they still resist classification capable of convincing a wide audience. For instance, some species which seem to arrange themselves into networks such as those members of the Rhizobiaceae, or Enterobacteriaceae, and others which may have clonal structures such as the Brucella organisms, represent difficult challenges for taxonomists.
The majority of plant pathologists and other professionals working with plants, consider a Rhizobium species one that is capable of nodulating root plants and fixing nitrogen, whereas an Agrobacterium species is capable of producing tumors in plants (after acquisition of Sym and Ti plasmids, respectively). During the last decade, prominent research has demonstrated that these bacteria are phylogenetically intermixed (Yanagi and Yamasato 1993), "promiscuous" with respect to the horizontal transfer of genes (Amábile-Cuevas and Chicurel 1992) and notably diverse (Laguerre et al. 1994, So et al. 1994), al! findings that have considerably complicated the classification of these organisms. Similar problems have been observed in the Salmonella and Pseudomonas species, whose diversity defeats the number of bacteriologists devoted to their study (LeMinor 1984, Palleroni 1984). It may be that the definition of species concerning these endosymbionts, plant pathogens, commensals, saprophytes and some animal pathogens must be viewed in the context of a reticular structure rather than as a unique terminal taxon (O'Hara 1994). Unfortunately, the undertaking of this laborious and difficult task does not guarantee that plant pathologists, soil microbiologists and medical doctors will accept the new nomenclature if it is of little practical use.
One has the tendency to think that the opposite would facilitate the classification of a bacterial species; however, this is not true. For instance, it is cIear that the Brucella species are a close related phylogenetic and taxonomic cluster of organisms (Fig. 1). As stated before, on the basis of DNA-DNA reassociation vaIues it has been proposed that all Brucella spp. are biovars of a single species, B. melitensis (Verger et al. 1985). Thus, this monospecies would inelude six biovars with their respective biotypes, e. g. B. abortus biotype 1 would become B. melitensis, biovar abortus, biotype 1. Even though this proposition may approach the suggestion of a "superior rank" of classification based on DNA-DNA hybridisation vaIues (Stackebrandt and Goebel 1994), the truth is that this molecular technique has only reveaIed genome similarities and no discreet, consistent and substantial differences that are known to exist among these organisms (Allardet-Servent et al. 1988, Meyer 1990, Fekete et al. 1992, Ficht et al. 1996). Although this may be one of the main reasons why this proposition has had little acceptance within the scientific community devoted to brucellae research (Meyer 1990), the ultimate judgement has come from veterinarians, microbiologists, physicians, epidemiologists and many other professionals who do not consider the new recommendation relevant for their purposes (Nielsen and Duncan 1990, Wong et al. 1992). The motive for this undoubtedly lies in the fact that for many years it has been known that B. melitensis is a "vicious bug" (once upon a time considered as one of the candidates for microbial warfare), while B. neotomae is a "n ice bug" confined to desert wood rats (Corbel 1989). This is not a matter of semantics; in every single country people responsible for control and eradication programs know the different pathological behaviour and epidemiological circumstances that distinguish the various Brucella species: talk of B. abortus is onething, but the mention of B. melitensis is a very different matter. The name of the latter species would dispatch a general alarm among all public health units (Flores-Castro and Baer 1979). Furthermore, tradition in many western countries indicates that polynomial names given to persons or to living things have a tendency to disappear in favour of (linnaean) binomial nomenclature. Therefore, just as one of the precursors of evolutionism, the "Conde Georges-Louis Leclerc de Buffon" is simply known as "Buffon", sooner or later "Brucella melitensis, biovar abortus, biotype 1", will probably be shortened to "B. abortus 1". In this respect it would be wise to weigh up the advantage of adopting a restrictive definition of species against well-established bacterial groups.
With respect to animals and plants it is accepted practice to define a new species on the basis of one or a few members or on the grounds of the discovery of a few fossil bones or seeds (Andersonand Jones-Jr 1984). Although some microbiologists have followed the same practice, in modern times very few scientists attempt to define a species on the basis of a single bacterial isolate. In general, the characterisation of repeated distinct isolates are required before a serious committee in bacterial nomenclature would consider it appropriate to define a new species (Graham et al. 1984). Nevertheless, this is not a straightforward subject. No one really knows how many isolates are necessary, and if these isolates should be obtained during certain time periods from different niches, habitats, hosts, localities and so on. In many cases the celerity for naming a new bacteria is based on the grounds of pure anthropocentric arguments. For instance in the case of some "unimportant" soil bacteria, endosymbionts or the pathogens of some marine animals there is no rush to provide a species name (EwaIt et al. 1994, Gao et al. 1994, So et al. 1994). However, when certain "emerging" pathogenic bacteria suddenly invade humans or animals and plants relevant for human use, the names of the new organisms are fiercely pursued (Regnery et al. 1992). Our human self-centred view of the world frequently makes us forget that bacteria themselves are not susceptible to systematics as we are.
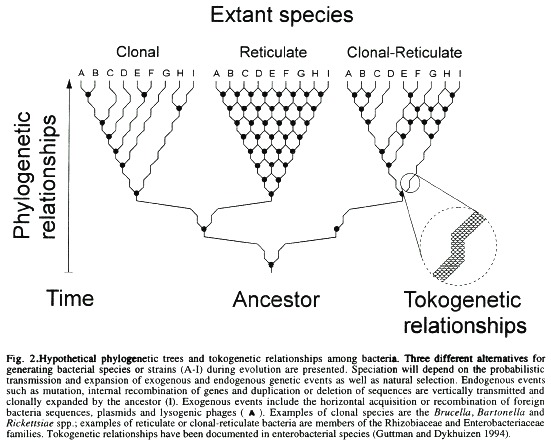
Are Bacterial Clonal or Reticulate Evolutionary Units?
Accessory genetic elements of bacterial genomes are plasmids, temperate phages, transposons, insertion sequences and retrons (Amábile-Cuevas and Chicurel 1992, Rice et al. 1993). These elements, besides being transmitted vertically during bacterial replication are also capable of horizontal transfer by means of conjugation, transformation, transduction, capsduction or retrotransposition. Under natural conditions the basic trend is that genes carried by accessory elements constitute a particular sample of the bacterial genome that is required occasionally rather than continually. By contrast, genes needed for normal functions located in chromosomes are the "housekeeping" stock that is indispensable (Krawiec and Riley 1990). However, there are a number of exceptions to these rules. Transposition and other mobilisation actions can shuttle specific genes and sequences between chromosomes and plasmids as well as group different genes into a single replicon (Amábile-Cuevas and Chicurel 1992). During this process some additional genes may be transferred by a particular phenomenon known as "DNA hitchhiking". The generalisation of this event has led to the suggestion that all bacteria may share a pool of genetic information that is accessible to virtually every other bacterial cell. Therefore, the bacterial community could be envisioned as a single, heterogeneous multicellular organism, with elements continually moving from one group of cells to another. In this respect, the bacterial universe would function as a superorganism with a network structure rather than as single entities with a branching family organisation (Sonea 1991). If this notion were assumed, the bacterial species concept would remain an illusion. Although this may sound exaggerated in the light of many different arguments given by population geneticists and evolutionary biologists who have convinced us that genetic identity is a reality in the microbial world (Ochman and Wilson 1987, Woese 1987, Olsen et al. 1994,), some bacterial species such as those belonging to the Rhizobiaceae and Enterobacteriaceae may assemble in a network fashion. In other cases, certain animal pathogenic bacterial species such as Bartonella spp. Brucella spp. Rickettsiae spp. and some endosymbionts seem to have a clonal structure because under certain local circumstances horizontal gene flux between bacteria, although possible, is an improbable natural event. From this perspective, the bacterial superorganism hypothesis, which is based on horizontal gene transfer between different species of bacteria, surrenders. Since these two standpoints (network vs. clonal structures) are important for a practical concept of bacterial species definition, some paragraphs will be devoted to their analysis.
For some time it was believed that recombination among bacteria occurred at such low frequency, relative to mutation, that it was considered evolutionary insignificant (Guttman and Dykhuizen 1994). However, recently it has been demonstrated that recombination is the dominant force driving the clonal divergence of some enterobacterial species (Guttman and Dykhuizen 1994), thus it must be considered a significant factor in the structuring of bacterial populations capable of horizontal gene transfer. Among enterobacterial species, recombination seems to take place more often between closely related bacteria than across "species". In this respect the genetic relationships between cJosely related individual s could be regarded as tokogenetic (Henning 1966) in contrast to the phylogenetic links that interconnect the different species. Tokogenetic recombination are expected to homogenise the gene pool among the interacting organisms, restricting the network structure of the species to a limited level (Fig. 2). In the case of some Escherichia coli strains, the divergence from a common ancestor was traced as recently as 2400 years ago (Guttman and Dykhuizen 1994). In comparison, recombination between different "species" (Amábile-Cuevas and Chicurel 1992) may result in genetic diversification promoting not only the expansion of the network structure of the species, but also favouring a fast and severe genetic drift which may eventually cause speciation (Syvanen 1994), in the manner proposed by the punctuated equilibrium hypothesis (Eldredge and Gould 1972). A similar recombinational phenomenon may explain the difficulties of defining bacterial species among certain groups of soil and plant associated bacteria (Laguerre et al. 1994, So et al. 1994).
The constant changes of the plantassociated bacteria from soil to plants would exert selective forces on the versatility of the genome, better achieved by non-essential genes carried in plasmids than in chromosomes (Eberhard 1990). However, the fact that chromosomal genes could move from one bacteria to another (Amábile-Cuevas and Chicurel 1992, Syvanen 1994), forces us to consider that some species of bacteria may arrange in a network structure. For instance members of the genera Agrobacterium and Rhizobium have been defined by their ability to induce plant tumors or root nodulation, respectively (Jordan 1984, Lambert et al. 1990). However, many non tumorigenic agrobacteriae and non nodulating rhizobiae have been isolated from soil (Bouzar et al. 1993, Laguerre et al. 1993, Kalogeraki and Winans 1995). In these bacteria the genes responsible for neoplastic transformation (onc and vir), root nodules (nod) and nitrogen fixation (fix and nif) are present in large accessory elements such as the Ti and Ri megaplasmids for gall and hairy root transformation respectively and the Sym megaplasmid for nodulation and nitrogen fixation (Jordan 1984, Lambert et al. 1990, Deng et al. 1995). The phylogeny of these plant-associated bacteria, based on ribosomal RNA sequences and supported by other genotypic characteristics (Dooley et al. 1993, Yanagi and Yamasato 1993, Wallington and Lund 1994), show that the species of these genera are intertwined (Fig. 1). Since Sym, Ti or Ri sequences are also intermingled with the different bacterial species, it is possible that the horizontal flux of these and other genes among the plant-associated bacteria has occurred several times during evolution (Deng et al. 1995). It is also probable that many of these genes located in plasmids or chromosomes have a common origin. This is further supported by the fact that Sym, Ti and R i plasmids share sequences among themselves as well as with genes located in the chromosomes (Prakosh and Schilperoort 1982, Kündig et al. 1993, Rivilla and Downie 1994, Schwedock and Long 1994, Kalogeraki and Winans 1995). Another important example that may sustain the network structure hypothesis of some species is the horizontal transfer of cytochrome oxidase genes between soil/plant bacteria. It has been found that a phylogenetic tree constructed on the basis of different cytochrome oxidase genes has unexpected assemblage with respect to phylogenetic trees constructed on the basis of ribosomal genes (Castresana et al. 1994). The incongruency seen between the cytochrome oxidase gene tree and the 16SrRNA tree suggests a case for horizontal gene transfer between these parties. Finally, several studies have demonstrated discrepancies between phylogenetic trees based on 16SrRNA and dendograms constructed on the basis of numerical taxonomy (Ladha and So 1994, Leung et al. 1994, So et al. 1994, Eardly et al. 1995). These findings support the notion that within this group of bacteria, the horizontal transfer of genes, rather than convergence or parallel evolution, bas influenced the evolution of species (Syvanen 1994). Interestingly, the high genetic diversity of plant-associated bacteria found worldwide, which overlaps with the clonal population structure encountered in specific ecosystems, is one of most striking characteristics of these organisms (Bouzar et al. 1993, Dooley et al. 1993, Leung et al. 1994, Wong et al. 1994).
As mentioned previously, there are a considerable number of intracellular bacteria such as Brucella, Bartonella, Rickellsiae, and Anaplasma species (Fig. 1) in which natural plasmids or temperate bacteriophages have not been detected in spite of numerous attempts to find them during chromosomal studies (Meyer 1990, Reschke et al. 1990, Regnery et al. 1991, Elzer et al. 1993, Roux and Raoult 1993, Grasseschi and Minnick 1994, Krueger et al. 1994). Furthermore, some of these bacteria, which have been experimentally transformed with conjugative plasmids, are incapable of transferring the extrachromosomal DNA to a second host of the same or a different species; and no evidence indicating that the plasmid had integrated into the chromosome was attained during these studies (Elzer et al. 1993). Gene transfer by other mechanisms such as translocation of naked DNA, conjugation mediated by specialised transposons, or reception of foreign DNA as result of heterologous conjugation cannot be ruced out. However, due to the characteristics of this bacteria) group, horizontal gene transfer seems an improbable natural event. In general terms, these intracellular organisms are distinguished by the following characteristics which are in accordance with the absence of foreign genetic material transference; 1) the metabolic and antibiotic susceptibility patterns are maintained constant throughout space and time; 2) structures such as pili, which favour the horizontal transference of genes between bacteria are absent in these pathogens ; 3) as a rule the animal pathogens are isolated as pure cultures, indicating that these bacteria are in less intimate contact with other species of bacteria; 4) infections by the animal pathogens are initiated by a few bacteria that are clonally expanded in the host; 5) multiple infections by different biovars seldom occur in the same host; 7) striking similarity among isolates of the same species obtained from different localities during different periods of time has been demonstrated; 8) bacterial diversity (e. g. number of species or biotypes) is commensurate with host and vector diversity (Ristic and Kreier 1984a, 1984b, Moulder 1985, lsenberg 1988, Corbel 1989, Halling and Zeher 1990, Meyer 1990, Regnery et al. 1991, Koehler et al. 1992, Schwartzman 1992, Brenner et al. 1993, Ewald 1993, Weiss and Moulder 1894, Birtles et al. 1995).
With respect to these intracellular animal pathogenic α-proteobacteria, and some bacterial endosymbionts (Yubin et al. 1994), the concept of species must be considered within the context of clonality rather than that of a network or reticulate structure. Since clonality is maintained by the vertical, asexual transmission of genetic material from parent to offspring, all evolutionary change through time must be considered the result of a mutational process or the autogenous rearrangement of genes mediated by insertion sequences, retrons or transposons. The relative contribution of mutation and autogenous recombination to divergence is not known. This is mainly due to problems associated with the collection of accurate figures during internal recombinational processes. However, in some plasmid-free bacteria such as Brucella spp. with an upper limit DNA-DNA reassociation values, the relative contribution between these two processes could be estimated by comparing the gene sequences at homologous sites in two different species, with the position of the genes in the chromosomes (Brucella spp. possesses two different chromosomes [Michaux-Charachon et al. 1993]). Differences in the number and location of insertion sequences between different Brucella species (Ouahrani et al. 1993), may suggest that autogenous recombination in addition to mutation could be a driving force in the speciation of bacteria devoid of heterologous recombination promoted by plasmids. It is possible that the speciation of intracellular bacteria and endosymbionts is commensurate with the evolutionary history of their hosts (Ochman and Wilson 1987). In this case the biological approach takes on an important dimension, since most of the environmental constraints that direct natural selection seem to be present in the host (Brunham et al. 1993, Laguerre et al. 1993). These interactions could attain extreme proportions such as mitochondria and chloroplasts in which the eubacterial identity is forever lost within the host environment, maintaining however, their phylogenetic relationship with the respective parental group (Yang et al. 1985). An important exception may be the modern and indiscriminate use of vaccines, antibiotics and other antimicrobial agents which may redirect some of tbe natural selective processes in these and other bacteria.
What is the Best Bacterial Species Definition?
For over a century, the biological species concept in the first instance, and following this, the taxonomic species concept were the major theoretical postulates orienting bacteriological research. In the 19th century and the first half of the 20th century bacteriology was primarily confined within the limits of medical research (Smith and Martin 1949). During this time many different pathogenic bacteria were described, and sets of key phenotype characters were established to identify members of the same. It was mainly in the second half of this century that research in bacteria devoid of medical importance began to participate in the microbiologist game. Owing to the incursion of molecular biology during the last decade, the bacterial world began to develop an evolutionary historical dimension which has greatly contributed to correcting some of the distorted perceptions "not in tune with the position of microorganisms in the natural order of things" (Woese 1987). The methods used for the definition of biological and taxonomic species concepts lack sufficient evolutionary perspective and, hence provide an inappropriate guide to the origins and products of evolutionary diversification (O'Hara 1993, Woese 1994). This has led to a call for the replacement of the biological-taxonomic concept by the phylogenetic species concept. However, as stated before, the methods used for constructing bacterial phylogenies do not in themselves possess sufficient resolution to distinguish those terminal taxons we call species; therefore a polyphasic phylogenetic-taxonomic approach was suggested (Stackebrandt and Goebel 1994). Although this latter proposition is closer to a practical definition of bacterial. species (or nomenspecies), it is vague with respect to the biological properties of the bacterial organisms. Furthermore, it does not take into consideration the microbiologist point of view, which I consider essential for a practical definition of bacterial species. As a scientist I am obligated to observe, measure, analyse and describe phenomena; however as a human being I may have preconceived notions of the object I am devoted to studying; this I can not neglect but if I recognise its existence, I may be able to control it.
The members of the α-2 proteobacteria shown in Fig. 1 which I have used to illustrate the virtues and drawbacks of the phylogenetic, taxonomic and biological approaches as well as the microbiologists perception, constitute a good exercise. For instance, it is clear that molecular sequencing based on 16SrRNA had resolved the phylogenetic position of facultative cell associated bacteria which were located haphazardly in different families (e.g. Rochalimae and Bartonella species), in the same family (e.g. Bradyrhizobium and Rhizobium species) or in groups with uncertain affiliation (e.g. Brucella and Ochrobactrum species). In addition, this approach has revealed that genera and the corresponding species of plant associated bacteria (e.g. Agrobacterium, Rhizobium and Phyllobacterium species) and some stem-nodulating and photosynthetic bacteria (Bradyrhizobium spp. and Rhodopseudomonas palustris) are intermixed in the phylogenetic tree, a fact that has called the attention of several investigators who have suggested reclassification of the group (Sawada et al. 1993, Willems and Collins 1993, Yanagi and Yamasato 1993). From a different perspective, the phylogenetic analysis did not have enough resolution in defining some of the animal (e.g. Brucella, Bartonella and Afipia species) and plant (Bradyrhizobium spp) associated species. In this regard numerical taxonomy has clearly resolved the position of some of the animal associated bacteria (Bartonella spp. and Afipia spp.) as demonstrated by the polyphasic approach undertaken to classify these organisms (Brenner et al. 1991, Brenner et al. 1993). However, while the data provided by 16SrRNA analysis gave a more reliable indication of the phylogeny, the degree of phenotypic divergence observed among some plant and soil bacteria (e.g. photosynthetic and non photosynthetic Bradyrhizobium spp.) and the degree of genotypic convergence estimated in Brucella spp. (Verger et al. 1985) raises questions concerning the polyphasic taxonomic approach to bacterial systematics (Yanagi and Yamasato 1993, So et al. 1994). Should these examples "persuade us that numerical taxonomy based on phenotypic characteristics does not provide reliable evidence concerning evolutionary relationships? Or should it instead remind us that sequence data alone do not tell us everything (or even anything) of interest about the biology of organisms" (So et al. 1994). At the present time both scenes may be correct. With respect to the soil and plant associated α-2 proteobacteria, the qualitative biological characteristics seem to be of little use in discerning different bacterial species; nevertheless it gives an excellent perception of the network structure of the group. Therefore it would be wise that microbiologists base their perception on this understanding, rather than on trying to dissect the soil and plant associated bacteria species on the grounds of 16SrRNA sequence comparisons and numeral taxonomy data alone. On the contrary, the phylogenetic data represented as trees as well as the DNA-DNA hybridisation analysis suggest to us that Brucella organisms are a monophyletic group of bacteria that may be clustered as a single species; although the qualitative biological attributes (e.g. different hosts and virulence) supported by fine analysis (AlIardet-Servent et al. 1988, Meyer 1990, Fekete et al. 1992, Ficht et al. 1996), clearly demonstrates that these bacteria behave as independent units deserving a distinct species classification. In this regard, it would be prudent that microbiologists base their perception on the cIonal structure exhibited by the Brucella organisms, rather than on the narrow classification displayed by the phylogenetic analysis and DNA-DNA hybridisation studies of these organisms (Meyer 1990).
A Never Ending Story
In considering a definition of bacterial species there is at least one major uncertainty left which probably would never be satisfactorily resolved. For instance, if bacteria behave as independent evolutionary units, adaptive changes may expand clonally resulting in a distinctive variety or in a new species. On the contrary, if they behave as reticulate evolutionary units, adaptive changes occurring within an individual are horizontally transferred to many or all members of the species which will "homogenise the species gene pool, even as it creates different genotypes and diversifies clones" (Guttman and Dykhuizen 1980). Recombination between different species may result in unbounded divergence and speciation. Depending on local circumstances bacteria may alternate from one form to another, that is, behaving as evolutionary units propagating clonally for long periods of time, followed by periods of "promiscuity" where clonal divergence and a network structure may result from recombination. Mutation without (foreign) recombination followed by cIonal expansion in an isolated environment (e.g. the intracellular milieu) may promote fast speciation, whereas recombination between cIones of the same species (tokogenetic relationships) may delay speciation. These ideas are schematically proposed in Fig. 2.
Without doubt, modem bacteriologists will devote much of their time to resolving these and other questions. Researchers will attempt to organise the knowledge of the diversity and variability among bacteria into systems of classification which reflect the evolution of these microorganisms as well as their similarities and differences under certain biological limits. Although this sounds relatively straightforward, the truth is that regardless of how much we learn about a particular bacterial group, the ambiguity related to the definition of species will prevail, because it is not possible to formulate "any clear distinction between individual differences and slight varieties; or between more plainly marked varieties and sub-species, and species" (Darwin 1859). No two individuals are exactly alike in all details. "Even if we were presented with a complete DNA sequence of every individual of every species, we would still not be able to discern what makes species different. We need to be able to modify genes to see if we can produce new strains and even new species, and see whether the species act as if they possess some inertia, resisting modification" (Statkin 1985). The diversity of living things is constantly emerging and chaotic; the historical nature of the evolution process impairs an inescapable indeterminate ness to the notion of species. This biological principle is comparable in many respects to the uncertainty principle of quantum mechanics proclaimed by Heisenberg more than 60 years ago, that established the impossibility of simultaneously deterrnining the velocity and position of a particle (Wheeler and Zurek 1983). The species problem is not a concept which will be solved, but rather something that we need to overcome, and the very existence of this uncertainty is itself one of the most significant pieces of evidence of the historical process we call evolution.
Acknowledgments
I thank Natasha McIver and several anonymous reviewers for their careful review of the manuscript. This work was supported by the Interfiiltional Atomic Energy Agency (8277/RB).
Resumen
Se examinó la definición de especie bacteriana usando como ejemplos protebacterias del subgrupo α-2 asociadas con las plantas y con los animales y tomando en consideración estudios filogenéticos. taxonómicos, biológicos y el criterio de los microbiólogos. La virtud que tienen los estudios filogenéticos es que proporcionan una perspectiva evolutiva de los linajes bacterianos; sin embargo el los métodos usados poseen poca resolución para definir aquellas especies localizadas en las ramas terminales de los árboles filogenéticos. El mérito que tienen los métodos taxonómicos se debe ha que definen las especies con base en múltiples características, permitiendo así una excelente resolución en las ramas terminales de los dendogramas; su desventaja radica en su poca exactitud en los nodos iniciales de los cladogramas. A nivel individual los estudios biológicos frecuentemente presenta dificultades debido a que muchas de las características son el resultado de coevolución, evolución paralela o bien transmisión horizontal de genes; sin embargo cuando este concepto es considerado conjuntamente con el filogenético y el taxonómico, se logran responder algunas preguntas que ninguna de las estrategias puede resolver aisladamente El juicio expresado por un grupo de expertos con base en evidencias científicas y en la práctica cotidiana para proponer una nueva especie de bacteria es lo que se conoce como el criterio de los microbiólogos. El éxito de este criterio se mide por su aceptación general y por su permanencia a través del tiempo. Uno de los problemas mas difíciles respecto a la definición de especie bacteriana es como distinguir si estas son unidades evolutivas independientes o si ellas son unidades evolutivas reticulares. En el primer caso, la herencia es trasmitida verticalmente como resultado de la división binaria y la multiplicación clonal. Este podría ser el caso de algunas bacterias asociadas con los animales en las que la recombinación genética posiblemente no ocurre o bien es excepcional. En el segundo caso, los cambios adaptativos que suceden en un individuo pueden ser trasmitidos horizontalmente a varios o a todos los miembros de un grupo. Este pareciera ser el caso de muchas de las bacterias intestinales y de las asociadas con las plantas. La divergencia genérica y la especiación de bacterias con una estructura clonal dependerán casi exclusivamente de mutaciones y de los rearreglos genéticos internos que puedan suceder, mientras que la especiación de bacterias con una estructura retícular dependerá no solo de estos procesos, sino también de las interacciones genéticas con otras bacterias. Esta incertidumbre, la que sin lugar a duda corresponde al proceso evolutivo, es al mismo tiempo uno de los elementos esenciales para comprender la definición de especie bacteriana.
References
Allardet-Servent, A. G. Bourg, M. Ramuz, M. Pages M. Bellis, & G. Roizes, 1988. DNA polymorphism in strains of the genus Brucella. J. Bacteriol. 170: 46034607. [ Links ]
Amáhile-Cuevas, C. F. & M. E. Chicurel. 1992. Bacterial plasmids and gene flux. Cell 70: 189-199. [ Links ]
Anderson, S. & J. K. Jones-Jr. 1984. Orders and families of recent mammals of the word. A Wiley-Interscience Publications, John Wiley & Sons, New York pp. 1-686. [ Links ]
Avise, J. C. & R. M. BaIl. 1990. PrincipIes of genealogical concordance in species concepts and biological taxonomy. Oxford Surv. Evol. Biol. 7: 45-67. [ Links ]
Avise, J. C. 1994. Molecular markers, natural history and evolution. Chapman & Hall Inc. , New York, London pp. 1-511. [ Links ]
Ayala, F. J. 1976. Molecular genetics and evolution, p.l120. In F. J. Ayala (ed.). Molecular evolution. Sinauer, Sunderland MA. [ Links ]
Birtles, R. J. T. G. Harrison, N. A. Saunders & D. H. Molyneux. 1995. Proposal to unify the genera Grahamella and Bartonella with description of Bartonella talpae combo nov. Bartonella peromysci combo nov. and the new species Bartonella grahamii sp. nov. Bartonella taylorii sp. nov. and Bartonella doshiae sp. nov. Int. J. Syst. Bacteriol. 45: 1.8. [ Links ]
Bouzar, H. D. Ouadah, Z. Krimi, J. B. Jones, M. Trovato. A. Petit & Y. Dessaux. Correlative association between resident plasmids and the host chromosome in a diverse Agrobacterium soil population. Appl. Environ. Microbiol. 59: 1310-1317. [ Links ]
Breitschwerdt, E. B. D. L. Kordick. D. E. Malarlcey, B. Keene, T. L. Hadfield & K. Wilson. 1994. Endocarditis in dogs due to infection with a novel Barlonella subspecies J. Clin. Microbiol. 33: 154-160. [ Links ]
Brenner, D. J. D. G. Hollis, C. W. Moss, C. K. English, G. S. Hall, J. Vincent, J. Radosevic, K. A. Birkness, W. F. Bibb. F. D. Quinn, B. Swaminathan. R. E. Weaver, M. W. Reeves, S. P. O'Connor, P. S. Hayes, F. C. Tenover, A. G. Steigerwalt, B. A. Perlcins, M. I. Daneshvar, B. C. HiII, J. A. Washington, T. C. Woods, S. B. Hunter, T. L. Hadfield, G. W. Ajello. A. F. Kaufmann. D. J. Wear & J. D. Wenger. 1991. Proposal of Afipia gen. nov. with Ajipia felis sp. nov. (formerly the cat scratcb disease bacillus), Ajipia clevelandensis sp. nov. (formerly the cleveland clinic foundation strain), Ajipia broomeae sp. nov. and Three Unnamed Genospecies, J. Clin. Microbiol. 29: 2450-2460. [ Links ]
Brenner, D. J. S. P. O'Connor, H. H. Winkler & A. G. Steigerwalt. 1993. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana combo nov. Bartonella vinsonii combo nov. Bartonella henselae combo nov. and Bartonella elizabethae combo nov. and to remove the family Bartonellaceae from the order Rickettsiales. Int. J. Syst. Bacteriol. 43: 777-786. [ Links ]
Brunham, R. C. F. A. Plummer & R. S. Stephens. 1993. Bacterial antigenic variation, host Immune response, and pathogen-host coevolution. Infect. Immun. 61: 2273-2276. [ Links ]
Castresana, J. M. Lübben, M. Saraste & D. G. Higgins. 1994. Evolution of cytochrome oxidase, an enzyme older than atmospeheric oxigen. EMBO J. 13: 2516-2525. [ Links ]
Corbel, M. J. 1989. Microbiology of the genus Brucella, p 53. In E. J. Young & M. J. Corbel (eds.). Brucellosis: chemical and laboratory aspects. CRC Press, Inc. Boca Raton, FL. [ Links ]
Cracraft, J. 1983. Species concept and speciation analysis, p.159-187. In R. F. Johoson (ed) Current ornithology, Plenum Press, NY. [ Links ]
Danto, A. 1985. Narration and knowledge. Columbia Univ. Press, New York. [ Links ]
Darwin, C. 1859. On the origen of species, p. 469. Murray, London. [ Links ]
Deng, W. Y. M. P. Gordon & E. W. Nester. 1995. Sequence and distribution of IS 1 312: evidence for horizontal DNA transfer from Rhizobium meliloli to Agrobaclerium lumefaciens. J. Bacteriol. 177: 2554-2559. [ Links ]
Dobzhansky, T. 1937. Genetics and the origin of species. Columbia University Press, New York. [ Links ]
Dooley, J. J. S. P. Harrison, L. R. Mynon, M. Dye. A. Cresswell, L. Skot, & J. R. Beeching. 1993. Phylogenetic grouping and identification of Rhizobium isolates on the basis of random amplified polymorphic DNA profiles. Can. J. Microbiol. 39: 665-673. [ Links ]
Dryden, S. C. & S. Kaplan. 1990. Localization and structural analysis of the ribosomal RNA operons of Rhodobacler sphaeroides Nucleic. Acids Res 18: 7267-7277. [ Links ]
Eardly, B. D. F. S. Wang, T. S. Whinam & R. K. Selander. 1995. Species Jimits in Rhizobium populations that nodulate the common bean (Phoseolus vulgaris). Appl. Environ. Microbiol. 61: 507-512. [ Links ]
Eberhard, W. G. 1990. Evolution in bacterial plamids and levels of selection. Quarterly Rev. Biol. 65: 3-22. [ Links ]
Eldredge, N. & S. J. Gould. 1972. Punctuated equilibria: an alternative to phyletic gradualism, p.82-1 15. In T. J. M. Schopf (ed.). Models in paleobiology, Freeman, Cooper & Co. San Francisco CA. [ Links ]
Elzer, P. H. M. E. Kovach, R. W. Phillips, G. T. Robertson; K. M. Peterson & R. M. Roop-II. 1995. In vivo and in vivo stability of broad-host-range cloning vector pBBiMCS in six Brucella species. Plasmid 33: 51-57. [ Links ]
Ewald, P.W. 1993. Evolution of Infectious Disease, Oxford Universiry Press Inc. New York. [ Links ]
Ewalt, D. R. J. B. Payeur, B. M. Martin, D. R. Cummins & W. G. Miller. 1994. Characteristics of a Brucella species from bonlenose dolphin (Tursiops Truncatus). J. Vet. Diag. Invest. 6: 448-452. [ Links ]
Fekete, A. 1. A. Bantle, S. M. Halling & R. W. Stich. 1992. Amplification fragment length polymorphism in Brucella strains by use of polymerase chain reaction with arbitrary primers. J. Bacteriol. 174: 7778-7783.
Ficht, T. A. H. A. Husseinen, J. Derr & W. Bearden. 1992. Species specific sequences at the omp2 locus for Brucella type strains. Int. J. Syst. Bact. 46: 329-331. [ Links ]
Flores-Castro, R. & G. M. Baer. 1979. Brucellosis (Brucella melilensis). Zoonotic implications, p. 195216. In J. H. Steele (ed.). Handbook series in zoonoses CRC Press, Inc. Boca Raton, FL. [ Links ]
Forterre, P. N. Benacbenhou-Lahfa, F. Confaloníeri, M. Duguet, C. EJie & B. Lahedan, 1993. The nature of the universal ancestor and the root of the tree of life. still open questions. Bio Systems 28: 15-32. [ Links ]
Gao, J. L. J. G. Sun, Y. Li, E. T. Wang & W. X. Chen, 1994. Numerical taxonomy and DNA relatedness of tropical rhizobia isolated from Hainan province, China. Int. J. Syst. Bacteriol. 44: 151-158. [ Links ]
Graham, P. H. M. J. Sadowsky, H. H. Keyser, Y. M. Barnet, R. S. Bradley, J. E. Cooper, D. J. De-Ley, B. D. W. Jarvis, E. B. Roslycky, B. W. Strijdom & J. P. W. Young. 1984. Proposed minimal standards for the description of new genera and species of root- and steam-nodulating bacteria. lnt. J. Syst. Bacteriol. 41: 582-587. [ Links ]
Grasseschi, H. A. & M. F. Minnick. 1994. Transformation of Bartonella bacilliformis by electroporation. Can. J. Microbiol. 40: 782-786. [ Links ]
Gray, G. C. A. A. Johnson, S. A. Thornton, W. A. Smith, J.' Knobloch, P. W. Kelly, L. O. Escudero. M. A. Huayd & R. L. Hoover. 1990. An epidemic of Oroya fever in Peruvian Andes. Ame. J. Trop. Med. Hyg. 42: 215. [ Links ]
Guttman, D. S. & D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination. not mutation. Science 266: 1380-1382. [ Links ]
Halling, S. M. & E. S. Zeher. 1990. Analysis of Brucella genotypic DNA for polymorphism and plasmids, p. 476. In L. G. Adams (ed.). Advances in brucellosls, Texas A&M University Press, College Station, TX. [ Links ]
Imhoff, J. F. & H. G. TrUper. Purple 1984. Purple nonsulfur bacteria (Rhodospirillaceae) Pfenning & TrUper 171. 17AL., p.1658. In N. R. Krieg & J. G. Holt (eds.). Bergey's manual of systernatic bacteriology, Vol 1. William & Wilkins, Baltimore. [ Links ]
Isenberg, H. D. 1988. Pathogenicity and virulence: another view. Clin. Microbiol. Rev. 1: 40-53. [ Links ]
Jordan, D. C. 1984. Family III. Rhizobiaceae Conn 1938. 321AL. p. 234. In N. R. Krieg & J. G. Holt (eds.), Bergey's manual of systematic bacteriology. Vol 1, WiIliam & Wilkins, Baltimore. [ Links ]
Kalogeraki. V. S. & S. C. Winans. 1995. The octapine-type Ti plasmid pTiA6 of Agrobacterium tumefaciens contains a gene homologous to the chromosomal virulence gene acvB. J. Bacteriol. 177: 892-897.
Koehler, J. E. Quinn. F. D. Berger. T. G. LeBoit, P. E. & J. W. Tappero. 1992. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. New Engl. J. Med. 327: 1625-1631. [ Links ]
Kordick. D. L. & E. B. Breitschwerdt. 1995. lntraerytbroytic presence of Bartonella henselae. J. Clin. Microbiol. 33: 1655-1656. [ Links ]
Krawiec, S. & M. Riley. 1990. Organization of bacterial chromosome. Microbiol. Rev. 54: 502.539. [ Links ]
Krieg. N. R. & J. G. Holt. 1984. Bergey's manual of systematic bacteriology, Vol 1, William & Wilkins, Baltimore. [ Links ]
Krueger. C. M. K. L. Marks & O. M. Ibler. 1994. Bartonella bacilliformis genome size estimate and preliminary macrorestriction map. Abstracts of th 94th general meeting, Ame. Soco Microbiol. H.187: 233. [ Links ]
Kumada, Y. D. R. Benson. D. Hillemann, T. J. Hosted. D. A. Rochefort, C. J. Tbompson. W. Woblleben & Y. Tateno. 1993. Evolution of the glutamine synthetase gene one of the oldest existing and functioning genes. Proc. Nat. Acad. Sci. 90: 3009-3013. [ Links ]
Kündig, C. H. Hennecke & M. Göttfert. 1993. Correlated physical and genetic map of the Bradyrhizobium japonicum 110 Genome. J. Bacteriol. 175: 613.622. [ Links ]
Ladha, J. K. & R. B. So. 1994. Numerical taxonorny of pbotosyntbetic nodulating Aeschynomene species. Int. J. Syst. Bacteriol. 44: 62-73. [ Links ]
Laguerre. G. M. AIlard. F. Revoy & N. Amarger, 1993. Isolation from soil of symbiotic and nonsymbiotic Rhizobium leguminosarum by DNA hybridization. Can, J. Microbiol. 39: 11 42.11 49 . [ Links ]
Laguerre, G. M. Bardin & N. Amarger. 1994. Rapic identification of Rbizobia by restriction fragment length polymorphism analysis of PCR-amplified 16SrRNA genes. Appl. Environ. Microbiol. 60: 56-63. [ Links ]
Lambert. B. H. Joos. S. Dierickx, R. Vantomme. J Swings, K. Kersters & M. Van-Montagu. 1990 Identification and plant interaction of PhyllobacJeriun sp. a predominant rhizobacterium of young sugar bee plants. Appl. Environ. Microbiol. 56: 1093.1102. [ Links ]
Le-Minor, L. 1984. Genus III. Salmanella Lingims 1900 389A1.. p. 427-458. In N. R. Krieg & J. G .Holt (eds.) Bergey's manual of systematic bacteriology. Vol I William & Wilkins, Baltimore.
Leung. K. S. R. Strain. F. J. DeBruijn & P. J. Bottomley 1994. Genotypic and phenotypic comparisons o cbromosomal types within an indigenous so; population of Rhizobium leguminosarum by. trifolii Appl. Environ. Microbiol. 60: 416-426. [ Links ]
Ludwing. W. J. Neumaier, N. KIugbauer. E. Brockmam: C. Roller, S. Jilg, K. Reetz. l. Schachtner. A Ludvigsen. M. Bachleitner, U. Fischer & K. H Schleifer. 1993. Phylogenetic relationships of bacteria based on comparative sequence analysis of elongation factor Tu and A TP-synthase beta subunit genes Antonie Van Leeuwenbock Int J. Gen. Mol. Biol 64: 3-4. [ Links ]
Meyer. M. E. 1990. Current concepts in the taxonomy of the genus BrucelIa. p. 1.17. In K. Nielsen &B. Donca (eds.). Animal brucellosis, CRC Press. Inc. BO( Raton, FL. [ Links ]
Michaux-Charachon, S. J. Paillisson, M. G. Carles-Nurit G. Bourg. A. Allardet-Servent & M. Ramuz. 1993 Presence of two independent chromosomes in the Brucella melitensis 16M genome. J. Bacteriol. 175: 701-705. [ Links ]
Moreno, E. 1992. BrucelIa evolution, p. 198-218. In Plommet (ed.). Prevention of brucellosis in mediterranean countries. International Centre for Advanced Mediterranean Agronomic Studies. Pudoc Scientific Publishers, Wageningen, Netherlands. [ Links ]
Moulder, J. W. 1985. Comparative biology of intracellular parasitismo Microbiol. Rev. 49: 298.337. [ Links ]
Nielsen, K. & B. Duncan. 1990. Animal brucellosis. CRC Press, Inc. Boca Raton, FL. [ Links ]
O'Hara. R. J. 1993. Systematic generalization historical fate, and the species problem. Syst. Biol. 42: 231-146. [ Links ]
O'Hara. R.J. 1994. Evolutionary history and the species problem. Am Zoologist 34: 12-22. [ Links ]
Ochman, H. & A. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in Cellular Genomes. J. Mol. Evol. 26: 74-86. [ Links ]
Olsen, G. J. C. Woese & R. Overbeek. 1994. the winds of (evolutionary) change: breathing new life into mirobiology. J. Bacteriol. 176: 1-6. [ Links ]
Olsen, GJ. & C.K Woese. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7: 113-123. [ Links ]
Ouahrani, S. S. Michaux. J. S. Widada. G. Bourg. R. Tournebize. M. Ramuz & J. P. Liautard. 1993. Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp. Relationship between genomic structure and number if IS6501 copies. J. Gen. Microbiol. 139: 3265-3273. [ Links ]
Palleroni, N. J. 1984. Genus I. Pseudomonas Migula 1894, 237 " (Nom. con. Opino 5, judo Comm. 1952,237), p. 141-199. In N. R. Krieg & J. G. Holt (eds.). Bergey's manual of systematic bacteriology, Vol 1. WiIliam & Wilkins, Baltimore. [ Links ]
Paterson. H. E. H. 1985. The recognition concept of species, p. 21-29. In E. S. Vrba (ed.). Transvaal Museum Monograph Nº 4, Pretoria, South Africa. [ Links ]
Pitt, T. L. 1994. Bacterial typing: the way ahead. J. Med. Microbiol. 40: 1-2. [ Links ]
Prakosh, R. K. & R. A. Schilperoort. 1982. The relationship between nif plasmids of fast growing Rhizobium species and Ti plasmids of Agrobacterium lumefaciens. J. Bacteriol. 149: 1129-1134. [ Links ]
Regnery, R. L. C. L Spruill,. & B. D. Plikaytis. 1991. Genotypic identification of Rickeltsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173: 1576-1589. [ Links ]
Regnery, K L. B. E. Anderson. J. E. Carridge-III, M. C. Rodriguez-Barradas, D. C. Jones & J. H. Carro 1992. Characterization of a novel Rochalimaea species, R. henselae sp. nov. isolated from blood of a febrile, human immunodeficiency virus-positive patient. J. Clin. Microbiol. 30: 265-274. [ Links ]
Reschke, D. K. M. E. Fraizer & L. P. Mallavia. 1990. Transformation of Rhochalimaea quintana, a member of the family Ricketlsiaceae. J. Bacteriol. 172: 51305134. [ Links ]
Rice, S. J. Bieber, J. Chung. G. Stacey & B. C. Lampson. 1993. Diversity of retrons elements of rhizobia and other gram-negative bacteria. J. Bacteriol. 175: 42504254. [ Links ]
Ristic, M. & J. P. Kreier. 1984a. Family II. Bartonellaceae Gieszcykiewiez 1939,25 AL p.717. In N. K Krieg & J. G. Holl (eds.). Bergey's manual of systematic bacteriology, Vol 1, William & Wilkins, Baltimore. [ Links ]
Ristic, M. & J. P. Kreier. 1984b. Family III. Anaplasmataceae Philip J957, 980 AL, p.719. In N. K Krieg & J. G. Holt (eds.). Bergey's manual of systematic bacteriology. Vol 1, WilIiam & Wilkins, Baltimore. [ Links ]
Rivilla. R. & J. A. Downie. 1994. Identification of Rhizobium leguminosarum gene homologous to nodT but located outside the symbiotic plasmid. Gene 144: 87-91. [ Links ]
Roux, V. & D. Raoult. 1993. Genotypic identification and phylogenetic analysis of the spotted fever group rickettsiae by pulsed-field gel electrophoresis. J. Bacteriol. 175: 4895-4904. [ Links ]
Saitou. N. & M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol 4: 406-425. [ Links ]
Sawada, H. H. leki, H. Oyaizu & S, Matsumoto. 1993. Proposal for rejection of Agrobacterium tumefaciens and revised description for the genus Agrobacterium and for Agrobacterium radiobacter and Agrobacterium rhizogenes. Int. J. Syst. Bacteriol. 43: 694-702. [ Links ]
Schadewald, R. 1986. Creationism pseudoscience p.305317. In K. Fraizier (ed.). Science confronts the paranormal, Prometheus books. NY. [ Links ]
Schwartzman. W. A. 1992. Infections due to Rochalimaea: The expanding clinical spectrum. Clin. Infect. Dis. 15: 893-902. [ Links ]
Schwedock. J. & S. R. Long. 1994. An open reading frame downstream of Rhizobium meliloti nod Q1 shows nucleotide sequence similarity to an Agrobacterium tumefaciens insertion sequence. Mol. Plant-Microbe Interact. 7: 151-153. [ Links ]
Simpson. G. G. 1951. The species concept. Evolution 5: 285-298. [ Links ]
Smith. D. T. & D. S. Martin. 1949. Zinsser's texbook of bacteriology. Appl.eton-Century-Crofts, Inc. New York, NY. [ Links ]
Sneath, P. H. A. 1984a. Numerical taxonomy. p. 5-7. In N. R. Krieg & J. G. Holt (eds.). Bergey's manual of systematic bacteriology. Vol 1, William & Wilkins, Baltimore. [ Links ]
Sneath. P. H. A. 1984b. Bacterial nomenclature. p.19-23. In N. R. Krieg & J. G. Holt (eds.). Bergey's manual of systematic bacteriology, Vol 1, William & Wilkins. Baltimore. [ Links ]
So, R. B. J. K. Ladha & J. P. W. Young. 1994. Photosynthetic symbionrs of Aeschynomene spp. form a cluster with Bradyrhizobia on the basis of fatty acid and rRNA analysis. lnt. J. Syst. Bacteriol. 44: 392-403. [ Links ]
Sonea, S. 1991. Bacterial evolution without speciation. p. 95. In L. Margulis & R. Fester (eds.). Symbiosis as a source of evolutionary innovation. MIT Press, Cambridge, MA. [ Links ]
Stackebrandt, E. & B. M. Goebel. 1994. A place for DNADNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44: 846-849. [ Links ]
Staley. J. T. & N. R. Krieg. 1984. Calssification of procaryotic organisms: an overview, p. 1-4. In N. R. Krieg & J. G. Holt (eds.). Bergey's manual of systematic bacteriology, Vol. 1, William & Wilkins, BaItimore. [ Links ]
Statkin, M. 1985. The descent of genes. Science 85, 6: 80-83. [ Links ]
Syvanen, M. 1994. Horizontal gene transfer: evidence and possible consequences. Ann. Rev. Genet. 28: 237- 261. [ Links ]
Templeton, A. R. 1989. The meaning of species and speciation: a genetic perspective, p.3-27. In D. Otte & J. A. Endler (eds.). Speciation and its consequences, Sinauet, Sunderland, MA. [ Links ]
Verger, J. F. Grimont, P. A. D. Grimont & M. Grayon. 1985. Brucella a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35: 292-295. [ Links ]
Viale, A. M. A. K. Arakaki, F. C. Soncini & R. G. Ferreyra. 1994. Evolutionary relationships among eubacterial groups as inferred from GroEL (chaperonin) sequence comparisons. Int. J. Syst. Bacteriol. 44: 527-533. [ Links ]
Wallington, E. J. & P. A. Lund. 1994. Rhizobium leguminosarum contains multiple chaperonin (cpn60) genes. Microbiology 140: 113-122. [ Links ]
Wayne, L. G. D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, P. Star & H. G. Troper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics Int J. Syst. Bacteriol. 37: 463-464. [ Links ]
Weisburg, W. G. M. E. Dobson, J. E. Samuel, G. A. Dasch, L. P. Mallavia, O. Baca, L. Mandelo, J. E. Sechrest, E. Weiss & C. R. Woese. 1989. Phylogenetic diversity of rickettsiae. J. Bacteriol. 171: 4202-4206. [ Links ]
Weiss, E. & J. W. Moulder. 1984. Family 1. Rickettsiaceae Pinkerton 1936 I 86 AL, p. 687-717. In N.R. Krieg & J. G. Holt (eds.). Bergey's manual of systematic bacteriology, Vol 1, William & Wilkins, Baltimore. [ Links ]
Wheeler, J. A. & W. H. Zurek. 1983. Quantum theory and measurement, Princenton University Press. [ Links ]
Willems, A. & M. D. Collins. 1993. Phylogenetic analysis of rhizobia and agrobacteria based on 16SrRNA gene sequences. Int. J. Syst. Bacteriol. 43: 305-313. [ Links ]
Williams, E. E. H. Rand, S. Rand & R. J. O'Hara. 1995. A computer approach to the comparison of species in difficult taxonomic groups. Breviora 502: 1-47. [ Links ]
Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51: 221-271. [ Links ]
Woese, C. R. 1994. There must be a prokaryote somewhere: microbiology's search for itself. Microbiol. Rev. 58: 1-9. [ Links ]
Wong, F. y. K. E. Stackebrandt, J. K. Ladha, D. E. Fleischman, R. A. Date & J. A. Fuerst. 1994. Phylogenetic analysis of Bradyrhizobium japonicum and photosynthetic stem-nodulating bacteria from Aeschynomene species grown in separated geographical regions Appl. Envíron. Microbiol. 60: 940-946. [ Links ]
Wong, J. D. J. M. Janda & P. S. Duffey, 1992. Preliminary studies on the use of carbon substrate utilization patterns for the identification of Brucella species. Diagn. Microbiol. Infect. Dis. 15: 109-113. [ Links ]
Yang, D. Y. Oyaizu, H. Oyaizu,G. J. Olsen & C. R. Woese, 1985. Mitochondrial origens. Proc. Natl. Acad. Sci. 82: 4443-4447. [ Links ]
Yanagi, M. & K. Yamasato. 1993. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16SrRNA gene using PCR and DNA sequencer. FEMS Microbiol. Letl. 107: 115-120. [ Links ]
Yubin, D. A. M. Dmitri & K. Chang. 1994. Monophyletic origen of a-division proteobacterial endosymbionts and their coevolution with insect trypanosomatid protozoa Blastocrithidia culicis and Crithidia spp. Proc. Natl. Acad. Sci. USA 91: 8437-8441 [ Links ]