Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.50 n.2 San José Jun. 2002
when feeding on different species of food plants?
Ana Sittenfeld 1, 3 , Lorena Uribe-Lorío 1 , Marielos Mora 1 , Vanesa Nielsen 1 , Glen Arrieta 1 and Daniel H. Janzen 2
1 Centro de Investigación en Biología Celular y Molecular (CIBCM), Universidad de Costa Rica, Ciudad Universitaria Rodrigo Facio, San José, Costa Rica.
2 Department of Biology, University of Pennsylvania, Philadelphia, PA 19104; djanzen@sas.upenn.edu
3 Corresponding autor. Tel.: +506 207 3204; Fax +506 207 3190; sitten@racsa.co.cr
Recibido 16-VII-2001. Corregido 21-I-2002. Aceptado 13-II-2002.
Abstract
We used classical culture techniques to explore gut bacteria and changes associated with dietary change in the highly polyphagous, tropical caterpillar Automeris zugana (Saturniidae). Fifty-five third instar wild-caught sibs feeding on Annona purpurea (Annonaceae) in the Área de Conservación Guanacaste (ACG) in northwestern Costa Rica were divided into eight groups. Each of seven groups was reared to the ultimate instar on another species of food plant normally used by A. zugana. Some pupae were also analyzed for the presence of bacteria. Aerobic bacterial cultures were obtained from all 33 caterpillar guts and the eight pupae inventoried. There was no clear pattern in species composition of cultivated bacteria among the eight diets, and each caterpillar on a given food plant carried only a small fraction of the total set of species isolated from the set of caterpillars feeding on that food plant. Taken as a whole, the larvae and pupae contained 22 species of cultivable bacteria in 12 genera. Enterobacter, present in 81.8% of the samples, was the genus most frequently isolated from the caterpillars, followed by Micrococcus and Bacillus. Bacillus thuringiensis was isolated from 30.3% of the dissected caterpillars, but found in caterpillars feeding on only half of the species of food plants.
Key words: Automeris zugana, Saturniidae, caterpillar, insect gut, bacterial diversity, food plant, Área de Conservación Guanacaste, Costa Rica.
Within a species-rich tropical biota of leaf-eating and foliage-inhabiting caterpillars (larvae of Lepidoptera), the great majority of species are monophagous or restricted in diet to a very few species of (usually) closely related food plants; this is certainly the case within the dry forest, cloud forest and rainforest ecosystems of the Área de Conservación Guanacaste (ACG) in northwestern Costa Rica (Janzen 1993, 2001, Burns and Janzen 2001, Janzen and Hallwachs 2001). However, there are a few species of ACG caterpillars that are relatively to extremely polyphagous. These generalists display two patterns of polyphagy (Janzen 1993, 2001). In one case, not discussed here, the species of caterpillar feeds on 5 - 10 closely related to quite unrelated species of plants in one ecosystem, but each individual caterpillar normally eats just one species of plant, the one that its mother laid the egg on (see Saturniidae and Sphingidae examples in Janzen 1985, 1993, 2001, Burns and Janzen 2001, Janzen and Hallwachs 2001). In the other case, an individual caterpillar feeds on 5 - 10s of species of plants, either by explicitly walking from plant to plant, or by fleeing the plant during a carnivores attack and not returning to that species of food plant. Such individual polyphagy normally occurs during the penultimate and ultimate larval instars (Janzen 2001). Since each caterpillar may chose to feed on different species of plants, the total fed on by that species may sum as high as 84 species in 26 families within a single ecosystem (Janzen 2001).
With some exceptions, little is known of the natural communities of microbes in the guts of insects (Santo Domingo et al. 1998). Recent studies on the hindgut of termites indicate that novel microorganisms from major groups of the domain Bacteria are specifically associated with this environment (Ohkuma and Kudo 1996, Paster et al. 1996, Kudo et al. 1998). A vast microbial diversity exists within one species of termite (Ohkuma and Kudo 1996), which speaks for a rich, relatively unexplored microbial diversity in Insecta. Essentially nothing is known about the normal gut microbiota of caterpillars in tropical environments or about microbiota changes associated with dietary change. Here we begin an exploration of the bacterial gut biology of one extremely polyphagous species of tropical caterpillar (Janzen 2001), that of the medium-large moth Automeris zugana (Saturniidae).
An unmolested A. zugana caterpillar normally grows from first to third instar on one species of food plant, one of the 5 - 10 species of plants on which the mother oviposited (Janzen 2001). Then in the penultimate and ultimate instar, it usually walks off this plant and feeds on other species. Alternatively, if molested by a carnivore in any instar, it is likely to drop off the plant to escape. It then commonly shifts onto other food plant species. Following such shifts in food plants, which are normal for this species of caterpillar, what happens to its gut microbiota?
We addressed this question by dividing up a large group of third instar sibs found in nature on one of the usual ovipositional food plants, Annona purpurea, and then rearing each small sib subgroup to the ultimate (fifth) instar on another species of "usual to rare" food plant. The bacteria of these caterpillar guts, and the early pupal stage, were explored through classical culturing techniques.
Materials and methods
Study site: The 55 sibling third instar A.zugana larvae were found on 06/27/1999 as a group (as usual for the early instars of this species) feeding in the A. purpurea crown in which their single egg clutch had been laid, in Bosque San Emilio of Sector Santa Rosa of the ACG (300 m elevation, 80 year old secondary successional forest). This date is six weeks after the six-month rainy season began. These caterpillars were part of the first caterpillar generation of the year of this bivoltine species.
Macroorganism natural history: Automeris zugana ranges from Costa Rica to Peru (Claude Lemaire 2000 pers. comm., Burns and Janzen 2001). Its food plant biology has been studied only in the ACG, and there primarily within the ACG dry forest ecosystem, where it is extremely common (Janzen 1993, Burns and Janzen 2001). The 1i100-plus rearing records for wild-caught A. zugana caterpillars in the ACG dry forest (Burns and Janzen 2001) reveal that penultimate and ultimate instar caterpillars may be encountered feeding on at least 84 species in 26 plant families, ranging from grasses to ferns, dicot herbs, shrubs, vines and enormous trees.
Automeris zugana caterpillars occur in habitats from the earliest stages of succession to deep old-growth forest understory, and in a great range of ecological conditions, from hot, dry and sunny open old fields to cool, moist and deeply shaded old-growth forest understory. They do, however, pass the dry and hot long dry season (January to late May) as a very desiccation-resistant pupa in a tough and tightly woven silk cocoon among the foliage or surface leaf litter.
In the ACG dry forest, an adult ecloses in the 2 - 4 week period following the beginning of the rains about mid-May, mates, lays strings of 70 - 120 eggs along the margins of one to several of at least ten species of (often unrelated) food plants over a 2 - 5 day period, and dies. The eggs use about 14 days to hatch, and the caterpillar feeds for 6 - 7 weeks, and spins a cocoon in the foliage. A second generation then occurs during the second half of the rainy season. At the end of the second generation, the pupae become dormant and remain that way to pass the six month dry season.
The first, second and third instar caterpillars are gregarious, feeding side-by-side until early penultimate instar unless the group is broken up by a carnivore attack. The penultimate and ultimate instars feed solitarily. The aposematic caterpillars (red and white pattern on light green) (Burns and Janzen 2001), are highly urticating and avoided by carnivores except those that either are specialists on Automeris caterpillars (most of their parasitoids) or specialists at catching urticating caterpillars (e.g., squirrel cuckoos, trogons). This life style –aposematic, slow-growing, individually protected, generalist– has been caricaturized in apposition to cryptic, faster-growing, often edible, specialist caterpillars (Janzen 1984).
Another trait of great relevance to the gut microbiota is that the A. zugana life style is also characterized by being a "leaf chipper" rather than a leaf chewer or "musher" (Bernays and Janzen 1984). That is to say, A. zugana does not chew a leaf but rather snips a piece off, as if with a pair of scissors, and swallows the piece without further mastication. The larger the caterpillar (the later the instar), the larger the piece of leaf. This is in contrast with many specialists (e.g., Sphingidae, Noctuidae, Notodontidae, Nymphalidae) that brutally crush and masticate the leaf fragment in the process of cutting it off, such that the ingesta is a puree (Bernays and Janzen 1984). The leaf chipper style of eating means that A. zugana gut microbes are living in a medium where many of the ingested nutrients are those that leak from a cut edge, and many of the plants defensive chemicals (especially the larger molecules such as resins and polyphenols) remain within the leaf. This contrasts strongly with the habitat for the microbes in a caterpillar gut where the ingesta is a finely milled puree in which all nutrients and all secondary defensive compounds are potentially fully accessible and exposed to active physiological processes and the microbes themselves.
Rearing conditions: Each caterpillar was confined to its own clear plastic bag that was previously unused and factory-fresh. Each bag was twisted shut and clothes-pinned to a line below a roof in a wall-less rearing barn at ambient temperature (see examples in photographs at http://janzen.sas.upenn.edu). The rearing barns are in the same dry forest in which the population of A. zugana breeds. The bags were exposed to indirect sunlight and rain splatter, but no direct sun or rain. A branchlet of the food plant with the caterpillar on it was placed in the bag. Food was changed each two days by taking out the branchlet and putting in a new freshly cut branchlet, but leaving in the bag the leaf on which the caterpillar was perched. Turds and frass in the bottom of the bag were left as is, and the bag was not inverted at the time of changing food. This is one of the standard methods by which hundreds of thousands of caterpillars have been reared in the ACG caterpillar inventory. It differs from the "natural" situation in that the caterpillar is free of carnivores, and that it must choose its food from among the leaves offered in the bag. The relative humidity in the plastic bag also approaches 100% continually, while in nature humidity fluctuates wildly with rain, wind, and nightfall.
Caterpillar food was collected by cutting off leafy branchlets of the desired species and placing them in a previously unused plastic bag in the forest (one plant species per bag) and then brought back to the rearing barn. These fresh branchlets were placed in the caterpillar bags within 1 - 2 hr of being cut. The foliage was not wet from recent rain at the time of collection. Foliage was not washed or manipulated in any other manner, and presumably was covered with the microbial array customary for that species of plant. The foliage collected of a given species of food came from 5 - 10 different individual plants during the course of the feeding trial, but all these plants were growing in the same secondary successional vegetation within 2 km of each other. The caterpillar pedigree was written on the outside of the plastic bag with a broad felt-tipped pen. This information was later captured into field notebooks at the time of caterpillar sacrifice.
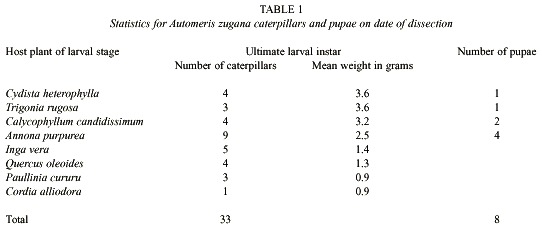
The experimental feeding lasted 20 – 21 days (June 28 to July 19, 1999) before the caterpillars were sacrificed for their gut contents 1 - 3 days before they would have become prepupae (they were 40 - 50 days old at this time). Thirteen larvae died of unknown causes (see discussion) during feeding. Nine larvae became prepupae and were allowed to pupate before dissection. The prepupa stops feeding, and defecates its gut contents, and then spins a cocoon. After several days of inactivity in the cocoon, the prepupa molts to the pupa. The 33 other larvae would have been prepupae if left to feed for 1 - 2 more days after the date of sacrifice. Caterpillar live weight and number of individuals available at the date of sacrifice varied with food plant (Table 1 and see discussion).
Food plants: The 55 sibling caterpillars were divided into groups of five to place on each of the eight food plant species (by accident six caterpillars were allocated to Calycophyllum candidissimum). Fourteen additional larvae were maintained on their natal food plant, Annona purpurea, so as to have specimens for additional experiments.
Seven of the eight species of plants fed to the caterpillars (Table 1) were chosen from among the 10 - 15 species on which A. zugana caterpillars are commonly found feeding in the ACG dry forest (Janzen 1984, 1985, 2001, Burns and Janzen 2001). Cordia alliodora, however, has been found as a food plant only ten out of more than 1i100 feeding records (Burns and Janzen 2001). Judging from the very low growth and survival rates of the caterpillars on C. alliodora, it is likely that these records represent the case of finding the caterpillar shortly before it moved on to other food plants.
There are no studies of how frequently an A. zugana caterpillar changes species of food plants on its own and when molested. The experiment conducted here represents just one of the many possible combinations of food plant experienced by the caterpillar, and represents the minimalist case of food diversity. The feeding pattern for an A. zugana caterpillar, free of molestation, is to pass the first through third instar feeding on the plant where its egg was laid, and then either at the end of the third instar, or early in the fourth instar (the penultimate instar) shift to a different species of food plant, where it may remain or later shift to yet others (Janzen 2001).
Sampling the caterpillar gut microbes: The last instar larvae were killed (1 - 3 days before they would have become prepupae) by placing them at -20ºC for 15 to 45 min depending on the size of the caterpillar. Prior to dissection, the caterpillar surface was disinfected by dipping the caterpillar in 70% ethanol, and it was rinsed several times with sterile distilled water to remove the ethanol. The same procedure was followed for the pupae. Each individual caterpillar was aseptically split open in a laminar flow cabinet at the ACG, and the intestinal tract removed. It was divided in three parts roughly corresponding to the foregut, midgut and hindgut. Each of these three samples was subdivided into three subsamples. One of the subsamples was placed in 70% ethanol for in situ hybridization and a second was placed in GIT buffer (Ogram 1998) for further DNA extraction reported elsewhere. The third was placed in nutrient broth, macerated and homogenized. A loop of this inoculated broth was immediately inoculated onto nutrient agar plates. Inoculated broth and plates were then transported at ambient temperature and aerobic conditions to the laboratories of the Centro de Investigación en Biología Celular y Molecular (CIBCM, Universidad de Costa Rica), located at a 5 hr drive from the ACG.
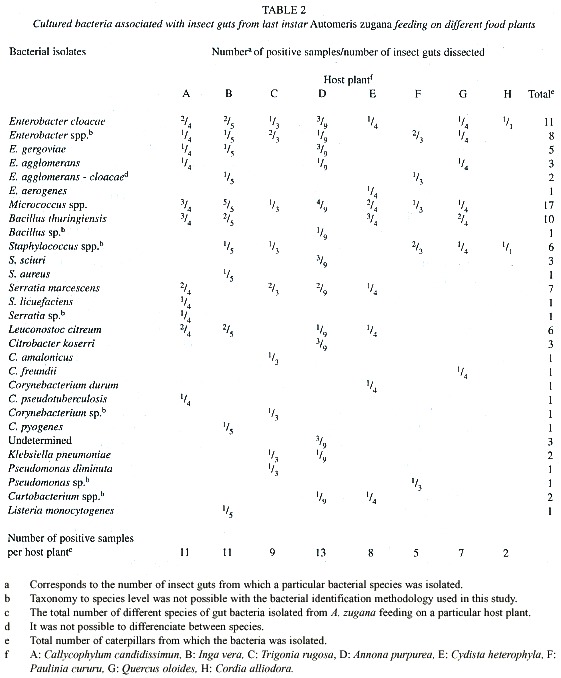
At the laboratory at CIBCM, the inoculated broth and plates were incubated for 48 hr at room temperature in aerobic conditions. This was followed by plating a loop of the inoculated nutrient broth in nutrient and Luria-Bertani (LB) agar, and incubated for one to four days at room temperature. The plates were surveyed for new colonies each 24 hr. Colonies were differentiated according to their morphology and through microscopic observation of their Gram stain traits. Representatives of each colony morphotype were picked from the plates. A single colony of each bacterial morphotype was chosen from representative isolates and transferred to a new plate. Cultures were considered to be pure after three successive passages on streak plates. Apparently different colonies were transferred to onto blood, nutrient and LB agar plates. One isolate of each type was then subjected to further biochemical tests and inoculated into various media for presumptive identification, using substrate utilization screening according to MacFaddin (1980) and Bergeys Manual of Determinative Bacteriology (Holt et al. 1994). API 20E strips from bioMérieux Vitek, Inc. Hazelwood, Mo. were used for the identification of some of the enteric bacteria. Tests for the ability of the isolates to grow on certain media were carried out in duplicate at room temperature and at 30ºC. All colonies growing in LB agar were selected and tested for Bacillus thuringiensis using the protocol described by Travers et al. (1987). Presumptive identification of cocci was based on gram staining, morphology, substrate use and biochemical tests. Several isolates could not be identified to genus level or to species level using the above methods. No bacterial counts were performed. The variable recorded was the number of different species of bacteria isolated from a given gut sample, rather than the total number of similar colonies observed in a plate (e.g. Tables 2 - 4).
Nine larvae that reached pre-pupal stages by July 20th , 1999, were left for 17 more days in the rearing barn in the ACG to develop into mature pupae. Eight of the nine pupae were dissected after washing the specimen for few seconds in 70% ethanol. Cocoons were carefully opened and portions of the dissected material (undifferentiated organs plus associated fluids) were directly transferred to tubes with either GIT or 70% ethanol for molecular analysis reported elsewhere. Other portions of the material were plated directly in nutrient and LB agar or added to nutrient broth and transported to CIBCM at room temperature.
At the CIBCM laboratories, the nutrient broths inoculated with pupal samples were incubated at room temperature for three days. The material was then plated on blood, LB and nutrient agar. LB plates were incubated for two days at 30ºC and tested for B. thuringiensis. Nutrient agar plates were incubated at room temperature to observe if colonies of other species were obtained after an enrichment in LB broth. The colonies that appeared on nutrient agar plates inoculated immediately from the dissected material showed only slow growth after 48 hr. These were then incubated for three more days at room temperature. Bacterial colonies were selected, transferred to blood, nutrient and LB agar, isolated to pure cultures, and identified as indicated above for those from the gut samples obtained from caterpillars.
Results
Food plants: The caterpillars survived and grew normally on seven of the eight food plants on which they were maintained during their fourth and fifth instars (Table 1). Cordia alliodora, however, is unambiguously a very marginal food plant. Not only did four of the five larvae given this food die slowly through what appeared to be "malnourishment" (they did consume large quantities of foliage), but the survivor was very small and would have produced a dwarfed moth, if it had survived. Equally notable was that this single surviving caterpillar had only two cultural species of bacteria in its gut samples (Table 2). Further discussion will be limited to the results with the seven species of food plants on which there was high survival and normal growth.
While survival was essentially the same on the seven "good" species of food plants, it is clear that the larvae did not add body weight or develop as fast on Inga vera, Quercus oleoides and Paulinia cururu as they did on Cydista heterophylla, Trigonia rugosa, Calycophyllum candidissimum and A. purpurea (Table 1). This is congruent with the observation in the field that these latter four species are the species on which last instar A. zugana larvae are most commonly encountered through casual search in the ACG dry forest and plants on which females commonly oviposit (DHJ). With the exception of I. vera, the guts of A. zugana feeding on these latter four plants had the greatest total diversity of culturable bacteria (Table 2). While the culturable bacteria from the guts of caterpillars fed I. vera were equally diverse, the caterpillars did not grow nearly as fast or as well as those on the other four mentioned above (Table 1).
Bacteria: Culturable bacteria were found in all the samples of caterpillar guts and pupal material inventoried (Tables 2 - 4). Since no differences were observed in the types of colonies from subsamples obtained from the different gut sectors (foregut, midgut and hindgut), the information from Tables 2 and 3, includes the total collection of isolates from all three sectors of an individual gut sample pooled as though it were a single sample.
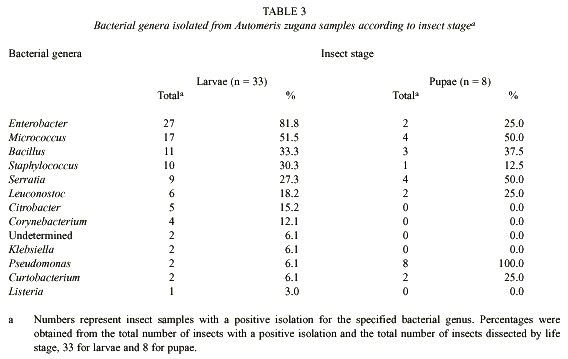
Considerable variation in bacterial genera was observed in different individuals. Bacteria identified are summarized in Tables 2 - 4. We identified a total of 19 different species of bacteria from 12 different genera from a total of 102 isolates (Table 2) from all A. zugana caterpillars. Thirty seven isolates were identified only to genus level and three could not even be identified to genus. These latter three were obtained from caterpillars feeding on A. purpurea. Two of the undetermined isolates were obtained from the same gut sample (Tables 2 and 3). Although these three isolates were different morphotypes of bacteria, it is not known whether they belong to the same genus (Table 3). The three unidentified isolates were gram negative rods, catalase positive, and oxidase negative. One showed oxidative metabolism, and the other two were fermentators that reacted differently on different substrates. One showed a positive reaction on methyl red, citrate and ornitine and was negative for Vogues Proskauer (VP). The other isolate was methyl red, urea and VP negative, utilized citrate and was able to grow on 15% NaCl.
There are differences in species composition of the gut microbiota in caterpillars feeding on different food plants (Table 2). In addition to the data shown in Table 2, we noted up to five different bacteria isolated from each caterpillar. The larvae feeding on the top five food plants for weight gains (Table 1) generated the most species of cultured bacteria (Table 2). Automeris zugana larvae feeding on leaves from A. purpurea generated the largest number of species of bacterial cultures (n = 13), followed by I. vera, C. candidissimum and T. rugosa. The same progression was observed for isolates from pupal samples. Larvae eating leaves from C. heterophylla, P. cururu, Q. oleoides, and C. alliodora produced lesser numbers of different bacterial samples, both in frequency per caterpillar and overall totals (Table 2).
Differences were observed in isolates from caterpillar guts and pupal material. At least 12 genera of bacteria were cultivated and identified from the caterpillar guts and pupal samples (Table 3). Taking into account that the undetermined genera (Tables 2 and 3) were gram negative, seven of the bacterial genera isolated were gram positive and at least seven were gram negative. The predominant genus isolated from the 33 caterpillars was Enterobacter, present in 81.8% of the samples (Table 2). Pseudomonas was present in all of the eight pupae studied. Klebsiella, Citrobacter, Listeria and Corynebacterium were not cultured from samples of pupae. The proportional representation of genera in the larval guts as compared with pupal guts were roughly the same except for the high frequency of Pseudomonas, and the very low frequency of Enterobacter, among the pupal samples (Table 3).
Not all the individuals fed the same plant species generated the same species of isolates. Caterpillars fed A. purpurea appeared to have the most similar bacterial flora among them. Only in one case did all five larvae feeding on one species of leaf generated a culture for the same species of bacteria (Micrococcus) and even in this case we are not convinced that there is only one species of Micrococcus (Table 2). Bacillus thuringiensis was isolated from 30.3% of the dissected caterpillars, but found in caterpillars feeding on only half of the species of food plants (Table 2).
We did not attempt to quantify the relative abundances of colonies appearing in the culture medium. However, it was obvious from the growth on the plates that Enterobacter, Serratia and Micrococcus were present at high density in the gut samples, and that no genera were present at high density in the pupal samples.
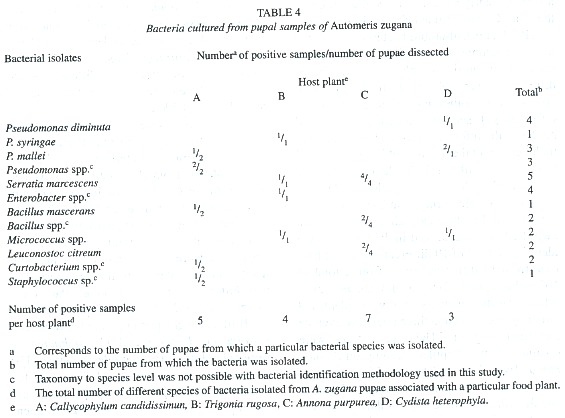
Bacterial isolates changed during development from caterpillar to pupa. Cultures from pupal material produced less abundant and less different colonies than did the samples from caterpillars (Table 2 and 4). It was common to observe just a few colonies representing each of the identified species isolated from pupal samples (results not shown). It was possible to isolate at least one colony from each individual pupa and a total of 30 different isolates in all samples combined. From these, 14 were identified only to genus level (Table 4). The largest number of different bacteria seven were cultured from pupae from caterpillars that had been feeding on A. purpurea. Serratia marcescens and representatives of the genus Pseudomonas were present in all four pupae associated with A. purpurea.
As mentioned earlier with respect to genus-level identifications, the cultured bacterial composition of the caterpillar guts and pupal material seems to differ when viewed at the species level (Table 2 and 4). Listeria, Corynebacterium, Klebsiella and Citrobacter were not isolated from pupae. At least 8 different genera of bacteria were culturable from A. zugana pupae samples (Table 3 and 4), and all were also found in the caterpillar gut samples. In six isolates it was not possible to classify the organisms to species level, but it was clear that the substrate utilization tests indicated differences from other species of the same genus identified in this study. It is also likely that yet other species of bacteria were present in the gut samples, but were not detected with the media used. Bacteria present in low numbers might also not have been detected with the culture techniques used in this study. There may well have been strict anaerobes and other viable but not culturable bacteria (Eutick et al. 1978, Felske et al. 1999).
Discussion
This initial inventory probe of the gut biota of A. zugana last instar larvae and pupae fed on different normal food plants demonstrates unambiguously that they are neither internally sterile nor occupied by a gut biota that is uniform among caterpillar individuals and among species of food plants. On the other hand, no clear pattern of food-plant-based differences in the culturable gut biota appeared, other than that those plants on which the caterpillars grew best generated the most species-rich sets of isolates.
Although there is recognition of the importance of microbial communities in insect guts, few studies have examined the composition of insect gut microbiotas. Early surveys of insect intestinal floras revealed the presence of many of the bacterial genera (and probably species) isolated in this work. In general, most of the bacterial genera and species reported here have been frequently isolated from animal intestines, including the guts of many termite species (Eutick et al. 1978), several species of mosquitoes (Demaio et al. 1996), the oil fly (Kadavi et al. 1999), and crickets (Ulrich et al. 1991). For example, Enterobacter, reported here as the most common bacterial genus, has also been reported as the major gut bacterium in termites belonging to the Rhinotermitiidae (Eutick et al. 1978), when aerobic culture techniques were used. Enterobacter and Citrobacter have also being suggested as important for fixing atmospheric nitrogen to produce a source of fixed nitrogen for the termites (Eutick et al. 1978, Janzen 1985). The significance of these findings to the dietary requirements of A. zugana is not known. Enterobacter cloacae, frequently isolated in our study, has being reported to survive and grow in the guts of silkworms (Watanabe et al. 2000).
It is interesting that B. thuringiensis was isolated from ten larvae (30% of the dissected caterpillars). This could have implications for the better understanding of ecological aspects of cry toxins in natural environments (Schnepf et al. 1998).
Based on other studies (Janzen 1985, Muyzer et al. 1993, Ohkuma and Kudo 1996, Paster et al. 1996, Liu et al. 1997, Kudo et al. 1998, Ohkuma et al. 1999), it is generally accepted that the culture approach fails to reflect much of the diversity present in insect guts. We assume that the species found cultured on our aerobic medium are only a small fraction of the species that were present, both as active populations and as "tourists" that were on the leaves when eaten. Our planned culture-independent approach, using molecular tools plus anaerobic and enrichment isolation techniques with the same caterpillars and pupae in this habitat (ongoing studies), will reveal just how unrepresentative are these inventory results using aerobic cultures on standard media.
Schultz and Breznak (1978), using strict anaerobic culture techniques to quantify heterotrophic bacteria in the hindguts of Reticulitermes flavipes, reported 66.3% streptococci among a total of 344 isolates. The results were obtained regardless of the origin of the termites, or the life stage or caste, or the duration of captivity. This suggests a very stable relationship between these bacteria and host. The remaining isolates were strains of Bacteroides and Enterobacteriaceae and a small percentage of strains of Lactobacillus, Fusobacterium and other anaerobic gram-positive rods. More recently (Ohkuma and Kudo 1996), using phylogenetic analysis of clonally isolated 16 S rDNAs, the intestinal microbiota of termites has been found to be highly diverse. Furthermore, two-thirds of the clones have less than 90% sequence identity to the known 16 S rDNAof cultivated organisms. Others (Ohkuma and Kudo 1996, Paster et al. 1996, Tokura et al. 2000) have reported Archeae and methanogenic bacteria in termite guts.
Assuming that the culture methods used were equally effective in their inventory of each sample, there is enormous heterogeneity among individual caterpillars. At the same time a wide variety of bacterial genera are present in caterpillar guts and pupae from A. zugana. Ecological interpretation of this result is not possible until it is understood whether 1) the cultured bacteria are merely tourists that happened to be inside the caterpillar (owing to having been on the leaves eaten), and/or 2) if they are a significant part of a gut biota that is made up of them plus the non-cultured biota that is undoubtedly present in the A. zugana gut. The alimentary tract of caterpillars is characterized by a very active epithelial transport, and the highest pH values known to be generated by a biological system (Dow 1992). The high pH, potential inhibitory enzymes (Russell and Dunn 1991), and defensive compounds from the plant foods, will affect the ability of caterpillar gut microbes to grow in culture.
It is striking that any given caterpillar on a given food plant carried only a small fraction of the total set of species isolated from the set of caterpillars feeding on that food plant species. Caterpillars fed A. purpurea appeared to have the most similar bacterial flora among them, but even here there are also quite strong differences among them. However, if the aerobically culturable species are only a tiny fraction of the total, the pattern in Tables 2 - 4 may be more accident than process. On the other hand, if the culturable microbiota is a large proportion of the total biota in a caterpillar, then this strong inter-caterpillar and inter-food- plant heterogeneity is both very puzzling and invites substantial exploration as to its cause and significance. In addition, it is simply not known what part of the caterpillar gut process is based on the raw nutrients leaked or masticated out of the leaf parts, and what part on microbe-driven processes. This brief inventory exploration shows, however, that there are many species of microbes present that could be either significant mutualists or merely "tourists" passing through.
Are sufficient bacterial raw materials present in different food plants to generate different gut biotas? The within-caterpillar and within-food plant heterogeneity in our results were so extreme that with the small samples sizes and the unknown thoroughness of the inventory methods, we cannot resolve this question. However, the data do not contradict the hypothesis that different caterpillars will have different gut biotas on different food plants. Santo Domingo et al. (1998), detected associated diet changes in cricket gut micro-biota, using hybridization techniques with group-specific probes. These diet changes altered the structure of the hindgut microbial community, although it did not change the predominance of eubacteria, as compared to the controls. These authors concluded that they could not demonstrate whether a structurally flexible hindgut microbial community could benefit the host.
It is tempting to view the bacteria within the A. zugana pupal material as the raw materials for vertical transmission of these microbes. While they certainly may be, they may also remain in the gut of the adult and not be part of maternal inoculation or contamination of eggs (the shells of which the newly hatched larvae eat). The reduction in the number of different bacteria between the larval stage and pupae, as well as the apparent reduction in the total number of isolates, is in agreement with Demaio et al. (1996). These authors, using quantitative aerobic bacterial cultures, revealed not only a complex midgut bacterial flora of wild-caught species of mosquitoes, with a total of 16 bacterial genera identified, but also differences in the microbial isolates from midguts during mosquito development and after blood-feeding. The authors suggested that the differences might be due to the presence of antibacterial compounds excreted into the gut during pupation, as has been reported for species of Lepidoptera and Diptera (Russell and Dunn 1991). Although the majority of microorganisms isolated by Demaio et al. (1996) were gram-negative rods, three gram-positive genera, including Bacillus, Brevibacterium and Enterococcus, were also isolated. Species most frequently isolated were Serratia marcescens, Klebsiella ozonae, Pseudomonas aeruginosa and Enterobacter agglomerans. Some of these species were also isolated from the A. zugana samples.
Studies on the house cricket (Acheta domestica) (Ulrich et al. 1991) indicated that part of the bacterial gut microbiota are not transient inhabitants of the gut lumen. Rather, they are attached to the peritrophic membranes in the midgut, the gut wall, and cuticular bristles in the hindgut. This was demonstrated by the failure to clear the wall of microbes when vigorously blended in a mixer (Ulrich et al. 1991). Of the 25 bacterial isolates obtained, 11 were identified as members of Citrobacter, Klebsiella and Yersinia (Ulrich et al. 1991). When comparing these results with the bacterial isolates from cockroaches, the authors indicate that although similarities exist in the diets and gut morphologies of crickets and cockroaches, each harbors distinctly different bacteria.
Caterpillar guts appear to be relatively uncomplicated straight tubular systems and some bacterial gut isolates might be transient. That we found only 22 different species of bacteria in 33 caterpillar guts and eight pupae, from eight different species of food plants, implies, but does not prove, that our culture isolates, are from resident populations rather than developed from the fortuitous tourist microbial species that just happened to be passing through. The isolates from pupae are even less likely to be simply contaminants from the caterpillar gut. Alternatively, if the species that we cultured are true residents, then there must be yet another "tourist" biota that will have to be revealed by a combination of DNA-based methods and/or by other kinds of culture methods, since leaves must have hundreds to thousands of species of microbes on them.
When the chipping generalist A. zugana caterpillar changes food plant, does its culturable gut microbiota change? Certainly the total set of species present in the caterpillars feeding on A. purpurea, the natal food plant for the caterpillars sampled in this study, is substantially smaller than the total found in the set of caterpillars feeding on the other seven food plants. In other words, the caterpillars did not simply retain whatever was their gut biota from A. purpurea when they were shifted to a new food plant. However, there is sufficient inter-caterpillar heterogeneity even on A. purpurea that the larger pattern displayed is simply enticing for further exploration rather than definitive.
Leaf-microbe ecosystems are poorly understood, but it is estimated that each leaf might be the habitat for at least one to ten million bacteria, and a number of factors affect the ecology of the phyllosphere microbiology at a given time (Hirano and Upper 2000). These factors exist in a continuously changing environment and include among others, plant and bacterial genetics, leaf surface and topography, plant chemistry, weather, and plant phenology (Hirano and Upper 2000). To this we would add the continual rain of fecal material from the animal kingdom as well as the debris from decomposition of all biodiversity. One of the few attempts to determine the structure and diversity of bacterial communities on leaves from a particular plant species revealed a large array of bacteria; Pseudomonas was the genus most frequently encountered (Hirano and Upper 2000).
Changes in the dominant microbial species on leaves must also occur across plant species and environmental conditions. There must be literally thousands of species of bacteria present on the foliage eaten by A. zugana. Of these, it must be possible to isolate many hundreds of bacterial species that were not considered in our study. On the other hand, plants and insects have coexisted for at least 100 million years, producing a great diversity of beneficial and negative relationships (Stotz et al. 1999, Dillon et al. 2000). The interaction of leaf microbe ecosystems and caterpillars adds another layer of complexity to the ecology of tropical ecosystems. The results of the present study, although preliminary, indicate fertile ground for research on the role of insect gut microbial communities in the interactions between the caterpillar, the microbial phylotypes consumed, and the caterpillar food plant.
Acknowledgments
The authors wish to express their gratitude to the staff of the ACG for providing logistics and support to the project. This work was supported in part by grant Ref 3-208-99 from CONICIT, San José, Costa Rica, by grant VI 801-99-506 from Vicerrectoría de Investigación, Universidad de Costa Rica, and facilitated by NSF DEB 9400829, DEB 9705072 to D.H. Janzen.
Resumen
Usamos técnicas de cultivo clásicas para explorar las bacterias del intestino y los cambios asociados con el cambio dietético en la, altamente polífaga, oruga tropical Automeris zugana (Saturniidae). Se capturaron 55 individuos hermanos, silvestres, de tercer estadio, alimentándose de Annona purpurea (Annonaceae) en el Área de Conservación Guanacaste (ACG) en el noroeste de Costa Rica y se dividieron en ocho grupos. Siete grupos fueron criados hasta el último estadio en otra especie de planta a la normalmente usada por A. zugana. Algunas pupas también fueron analizadas para ver la presencia de bacterias. Se obtuvieron cultivos de bacterias aeróbicas de los 33 intestinos de las orugas y de las ocho pupas muestreadas. No hubo un patrón claro en la composición de especies de las bacterias cultivadas entre las ocho dietas y cada oruga en una planta dada representó solo una pequeña fracción del conjunto total de especies aisladas de orugas alimentándose de esa planta. Tomándolas como un solo grupo, las larvas y pupas tuvieron 22 especies de bacterias cultivables en 12 géneros. Enterobacter, presente en 81.8% de las muestras, fue el género aislado de las orugas con más frecuencia, seguido por Micrococcus y Bacillus. Bacillus thuringiensis fue aislado de 30.3% de las orugas disectadas, pero encontrado en orugas alimentándose de solo la mitad de las especies de plantas.
References
Bernays, E.A. & D.H. Janzen. 1988. Saturniid and sphingid caterpillars: Two ways to eat leaves. Ecology 69: 1153-1160. [ Links ]
Burns, J.M. & D.H. Janzen. 2001. Biodiversity of phyrrhopygine skipper butterflies (Hesperiidae) in the Área de Conservación Guanacaste, Costa Rica. J. Lep. Soc. (in press). [ Links ]
Demaio, J., C.B. Pumpuni, M. Kent & J.C. Beier. 1996. The midgut bacterial flora of wild Aedes triseriatus, Culex pipiens and Psorophora columbiae mosquitoes. Amer. J. Trop. Med. Hyg. 54: 219-223. [ Links ]
Dillon, R.J., C.T. Vennard & A. Keith Charnley. 2000. Exploitation of gut bacteria in the locust. Nature 403: 851. [ Links ]
Dow, J.A. 1992. pH gradients in lepidopteran midgut. J. Exp. Biol. 172: 355-375. [ Links ]
Eutick, M.L., R.W. OBrien & M. Slaytor. 1978. Bacteria from the gut of Australian termites. Appl. Environ. Microbiol. 35: 823-828. [ Links ]
Felske, A., A. Wolterink, R. van Lis, W.M. de Vos & A.D.L. Akkermans. 1999. Searching for predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30: 137-145. [ Links ]
Hirano, S. & C.D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae -a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64: 624-653. [ Links ]
Holt, J.G., N.R. Krieg, P.H.A. Sneath, J.T. Staley & S.T. Williams. 1994. Bergeys manual of determinative bacteriology. Williams and Wilkins, Baltimore. [ Links ]
Janzen, D.H. 1984. Two ways to be a tropical big moth: Santa Rosa saturniids and sphingids. Oxford Surv. Evolut. Biol. 1: 85-140. [ Links ]
Janzen, D.H. 1985. A host plant is more than its chemistry. Illinois Natur. Hist. Bull. 33: 141-174. [ Links ]
Janzen, D.H. 1993. Caterpillar seasonality in a Costa Rican dry forest, p. 448-477. In N.E. Stamp & T.E. Casey (eds.). Caterpillars. Ecological and evolutionary constraints on foraging. Chapman and Hall, New York. [ Links ]
Janzen, D.H. 2001. How polyphagous are Costa Rican dry forest saturniid moth caterpillars? In Y. Basset, V. Novotny, S.E. Miller & R. Kitching (eds.). Arthropods of tropical forests: Spatio-temporal dynamics and resource use in the canopy. Cambridge University, Cambridge. (In press.) [ Links ]
Kadavy, D.R., B. Plantz, C.A. Shaw, T.A. Myatt, T.A. Kokjohn & K.W. Nickerson. 1999. Microbiology of the oil fly, Helaeomyia petrolei. Appl. Environ. Microbiol. 65: 1477-1482. [ Links ]
Kudo, T., M. Ohkuma, S. Moriya & K. Ohtoko. 1998. Molecular phylogenetic identification of the intestinal anaerobic microbial community in the hindgut of the termite, Reticulitermes speratus, without cultivation. Extremophiles 2: 155-161. [ Links ]
Liu, W.T., T.L. Marsh, H. Cheng & L.J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63: 4516-4522. [ Links ]
MacFadin, J.F. 1980. Biochemical test for identification of medical bacteria. Williams and Wilkins, Baltimore. [ Links ]
Muyzer, G., E.C. De Waal & A.G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polimerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ Microbiol. 59: 695-700. [ Links ]
Ohkuma, M. & T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62: 461-468. [ Links ]
Ohkuma, M., S. Noda & T. Kudo. 1999. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl. Environ Microbiol. 65: 4926-4934. [ Links ]
Ogram, A. 1998. Isolation of nucleic acids from environmental samples, p. 273-334. In R.S. Burlage, R. Atlas, D. Stahl, G. Geesey & G. Sayler (eds.). Techniques in microbial ecology. Oxford University, New York. [ Links ]
Paster, B.J., F.E. Dewhirst, S.M. Cooke, V. Fussing, L.K. Poulsen & J.A. Breznak. 1996. Phylogeny of not-yet-cultured Spirochetes from termite guts. Appl. Environ. Microbiol. 62: 347-352. [ Links ]
Russell, V.W. & P.E. Dunn. 1991. Lysosyme in the midgut of Manduca sexta during metamorphosis. Arch. Insect. Biochem. Physiol. 17: 67-80. [ Links ]
Santo Domingo, J.W., M.G. Kaufman, M.J. Klug & J.M. Tiedje. 1998. Characterization of the cricket hindgut microbiota with fluorescently labeled rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64: 752-755. [ Links ]
Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D.R. Zeigler & D.H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62: 775-806. [ Links ]
Schultz, J.E. & J.A. Breznak. 1978. Heterotrophic bacteria present in hindguts of wood-eating termites [Reticulitermes flavipes (Kollar)]. Appl. Environ. Microbiol. 35: 930-936. [ Links ]
Stotz, H.U., J. Kroymann & T. Mitchell-Olds. 1999. Plant-insect interactions. Curr. Opin. Plant Biol. 2: 268-272. [ Links ]
Tokura, M., M. Ohkuma & T. Kudo. 2000. Molecular phylogeny of methanogens associated with flagellated protists in the gut and with the gut epithelium of termites. FEMS Microbiol. Ecol. 33: 233-240. [ Links ]
Travers, R., P.A. Martin & C.F. Reicheldelfer. 1987. Selective process for efficient isolation of soil Bacillus spp. Appl. Environ. Microbiol. 53: 1263-1266. [ Links ]
Ulrich, R.G., D.A. Buthala & M.J. Klug. 1981. Microbiota associated with the gastrointestinal tract of the common house cricket, Acheta domestica. Appl. Environ. Microbiol. 41: 246-254. [ Links ]
Watanabe, K., K. Abe & M. Sato. 2000. Biological control of an insect pest by gut-colonizing Enterobacter cloacae transformed with ice nucleation gene. J. Appl. Microbiol. 88: 90-97. [ Links ]
Internet references
Janzen, D.H. & W. Hallwachs. 2001. Philosophy, navigation and use of a dynamic database ("ACG Caterpillars SRNP") for an inventory of the macro-caterpillar fauna, and its food plants and parasitoids, of the Área de Conservación Guanacaste (ACG), northwestern Costa Rica. (Downloaded: Feb., 12, 2002. http://janzen.sas.upenn.edu). [ Links ]