Cellulose, the main constituent of plant cell walls, is the world’s most abundant polysaccharide and with an annual production estimated at 1.5 x 1012 tons of biomass (Klemm, Heublein, Fink, & Bohn, 2005). Dry biomass on Earth, mainly cellulose, is estimated to 1.85 - 2.4 x 1012 tons (Hodásová, Jablonský, Škulcová, & Ház, 2015). This represents an almost inexhaustible source of polysaccharides, which may be harnessed for industrial and biotechnological purposes. Brazil is the largest producer of ethanol from sugar cane, a process that generates lignocellulosic residues of around 250 kg of bagasse for each liter of ethanol (Balat, 2011). While this residue can be converted to biofuel, the resistance of lignocellulosic materials hampers its direct bioconversion in fermentative processes (Sticklen, 2008).
Degradation of lignocellulose into sugars is the first step required for its use and can be achieved chemically, enzymatically or by a combination of both processes (Balat, 2011). Cellulase (EC 3.2.1.4), laccase (EC 1.11.1.7) and lipase (EC 3.1.1.3) are three enzymes involved in the degradation of lignocellulose. Lipase acts as a catalyst of this process, breaking down acids and resin formed during lignocellulose degradation (Karlsson, Holmbom, Spetz, Mustranta, & Buchert, 2001).
Fungi are important producers of enzymes for industry, with cellulase accounting for nearly 20 % of the total world enzyme market (Lange et al., 2012). They are key players in litter decomposition, due to the fact they produce a wide range of extracellular enzymes and are resistant to environmental stress (Valencia & Chambergo, 2013). They are therefore of interest for biotechnological applications in food, wine and textiles industries (Bhat, 2000; Viswanath, Rajesh, Janardhan, Kumar, & Narasimha, 2014), for energy generation (Balat, 2011), for plastics degradation (Kathiresan, 2003) and to produce high-added value compounds such as biosensors, cosmetic products and organic acids (Hasan, Shah, & Hameed, 2006; Oliveira & Graaff, 2011). Bioprospecting microorganisms that can produce enzymes with higher specific activity and efficiency are mandatory in biotechnological processes.
It is estimated that there are approximately 5.1 million fungi species in the world, of which only 2 % are known (Blackwell, 2011; Maia et al., 2015). While the Brazilian savanna is a biodiversity hotspot, it is subject to a high rate of deforestation (Trancoso, Sano, & Meneses, 2015) which decreases fungal diversity in its soil (Castro et al., 2008). The soil of the region is red or yellow latosol, has a low pH, low cation exchange capacity, high aluminum saturation and a high clay content (Oliveira-Filho & Ratter, 2002; Castro et al., 2008; Castro, Silva, Quirino, Bustamante, & Krüger, 2016). There is a great number of non-identified microorganisms in this environment (Castro et al., 2008; Vieira, Johann, Hughes, Rosa, & Rosa, 2014; Maia et al., 2015; Castro et al., 2016) and, since high aluminum saturation is generated due to laccase enzymatic activity (Viswanath et al., 2014), this feature indicates Cerrado soil might harbor lignocellulolytic fungi. Leaf litter is also an important source of cellulolytic fungi, as they produce extracellular enzymes to degrade wood and leaves (Korkama-Rajala, Mueller, & Pennanen, 2008).
The aim of the present study, therefore, was to isolate, identify and detect cellulase and lipase activity in fungi collected from the Cerrado State Park in Brazil, and in this way, to provide further information on the biotechnological resources of the Brazilian savanna.
Material and methods
Area: The soil and leaf litter samples were collected in the Cerrado State Park, located in the town of Jaguariaíva (24º09’S & 50º18’W) and which has a total area of 2 256.62 ha (Figure 1). The Köppen-Geiger climate classification is Cfb (Alvares, Stape, Sentelhas, Gonçalves, & Sparovek, 2013). Sampling was performed only once, at the beginning of rainy season on September, 2014. The climatic conditions were temperature 22 °C, humidity 66 %, a wind speed of 9 km/hand light rain. Sampling was performed at three random points of the park: 24°11’229”S - 49°39’997”W; 24°11’051”S - 49°39,992’W and 24°10’524”S - 49°40’085”W (Figure 1, inset) only once. Vegetation of the three points were similar, and consisted of xeromorphic vegetation, with woody trees and shrubs scattered over grassland, sometimes forming a dense bush (Linsingen, Sonehara, Uhlmann, & Cervi, 2006). Two samples from each point were taken, being one of leaf litter and the other one of soil. Samples of leaf litter were collected and then this layer was removed. Subsequently, soil from the same point was sampled to a depth of 10 cm below the surface. The samples were stored in sterile plastic bags and transported at room temperature to the Environmental Microbiology Laboratory (UFSCar, Sorocaba, SP, Brazil) and kept at 4 oC for three days until the beginning of the analysis.
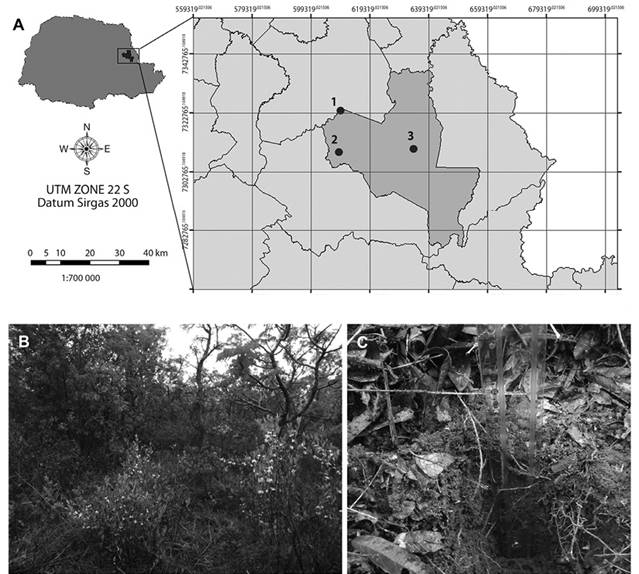
Figure 1 A) Location of Cerrado State Park in Paraná state, Brazil. Inset shows location of sampling points inside the park (modified from www.itcg.pr.gov.br). B) Brazilian savanna vegetation in Cerrado State Park, point 1. C) Soil sampling below leaf litter cover. Plastic ruler marks sample depth. Vegetation and leaf litter cover was similar in the three sampling points.
Physicochemical analysis: The pH of the soil and leaf litter samples was measured using pH-FIX 0-14 litmus paper tape (Macherey-Nagel, REF 92110). The soil particle size test was performed according to previously established parameters using sieved fractions (Camargo, Moniz, Jorge, & Valadares, 2009).
Isolation and identification of filamentous fungi: Samples were weighed into three portions of 1 g each. Leaf litter samples were macerated in a mortar. Soil and macerated leaf litter samples were diluted in 9 mL of 0.9 % saline solution and serial diluted up to 10-5, without replicas. These dilutions were inoculated (0.1 mL/L) in duplicate in medium with cellulose, adapted from Parkinson, Gray and Williams (1971), L-asparagine was substituted for peptone and 500 mg/L ampicillin (Hankin & Anagnostakis, 1975) was added. The plates were subsequently incubated at 30 °C for five days. Selected grown colonies were transferred to potato-dextrose-agar (PDA 20 g/L) plates. Colony characteristics such as growth, shape and color were observed. Filamentous fungi were isolated by successive inoculations in PDA maintained at 30 °C.
DNA extraction from mycelia grown in PDA for seven days were performed in accordance with Silva, Bernardi, Schaker, Menegotto, & Valente, (2012). Partial 18S rRNA gene was amplified using the primers NL1 (5’GCATATCAATAAGCGGAGGAAAAG-3’) and NL4 (5’-GGTCCGTGTTTCAAGACGG-3’) (Lopes, Soden, Martens, Henschke, & Landridge, 1998). Amplification consisted of 30 cycles of denaturation at 94 °C for 45 s, annealing at 50 °C for 1.5 min, and extension at 72 °C for 1.5 min. An initial 4 min denaturation at 94 °C and a final 7 min extension at 72 °C were performed. Amplified products were purified using Illustra GFX PCR DNA and Gel Band purification (GE Healthcare), and sequenced using a ABI PRISM 3730 DNA Analyzer (Applied Biosystems, Foster City, California, USA) with Big Dye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems). The BLASTn tool from the National Center for Information Biotechnology was used to compare the sequences obtained with those deposited in GenBank (Benson et al., 2013). Sequences were deposited in GenBank under accession numbers KY616857 to KY616884.
Cellulase activity: The ability of the isolates to produce cellulose was tested by placing a loopful of fungi of each isolate into the center of an agar plate containing 10 g/L carboxymethyl cellulose (CMC). The composition of the medium was (g/L): CMC (10) and agar (10). Each plate was inoculated in duplicate and maintained for seven days at 30 °C. The production of cellulose was verified by applying 10 mL of lugol over the colonies, and CMC-ase-positive strains were identified by the clear zones surrounding individual isolates (Kasana, Salwan, Dhar, Dutt, & Gulati, 2008). Pleurotus sp. was used as the positive control, due to its high cellulose activity (Mandeel, Al-Laith, & Mohamed, 2005).
Lipase activity: Lipolytic activity was assessed using the medium described by Singh, Gupta, Goswami, and Gupta (2006), but with fungi strains inoculated on test plates containing phenol red and tween 20. Plates were incubated at 37 °C for four days. The formation of lipolytic enzymes was verified by pH variation. The medium with Rhodamine B (Kouker & Jaeger, 1987) was used to fluorimetrically detect the production of lipase. The plates were incubated at 30 °C for six days. Rhodamine B reacts with fatty acids and when placed under UV light, it emitted fluorescent radiation around the colonies, however, the size of the halo was too small to be measured and results were read by the emission (positive) or no emission (negative) of fluorescence.
Results
A total of 28 strains of filamentous fungi were isolated from soil and leaf litter samples in CMC medium (Table 1). Most strains belonged to the Ascomycota phylum (83 %) and 17 % belonged to the Basidiomycota phylum. The soil samples differed in texture and pH but presented a similar leaf litter cover (Figure 1). Most isolated strains in all samples belonged to the Ascomycota phylum. The four isolated fungi from clay soil samples collected in area 1, with pH 6, belonged to the Ascomycota phylum and two strains were identified as belonging to the order Pleosporales, one of them identified as genera Bipolaris sp., and the species Penicillium funiculosum and Colletotrichum boninense. Four Ascomycota and one Basidiomycota fungi originated from the soil samples collected in point 2 (sandy clay, pH 5), and were identified as the genera Curvularia sp., Trichoderma sp., Bipolaris sp., Bjerkandera sp. and the species Penicillium funiculosum and Penicillium camemberti. Soil sample 3 (sandy, pH 4) generated four Ascomycota and one Basidiomycota fungi, including the genera Penicillium sp. and the species Alternaria alternata, Bjerkandera adusta and Curvularia lunata (Table 1). The three leaf litter samples presented pH 7, however, different fungi were isolated from each sample. Only two Basidiomycota strains were isolated from leaf litter, Trametes sp. from leaf litter 2 and Bjerkandera sp. from leaflitter 3. Trichoderma sp. was isolated from the three samples, while Talaromyces sp. and Penicillium sp. were isolated from samples 1 and 3. Fusariumsp., Hirsutella sp. and Bipolaris sp. were isolated only once, from samples 1, 2 and 3, respectively (Table 1).
Table 1: Identification of fungi isolated from leaf litter and different soil types from Brazilian Savanna
| Sample | Taxa | % Iden. | % cover | E value | Access number |
|---|---|---|---|---|---|
| Soil 1 | Pleosporales | 100 | 100 | 0 | HE820842.1 |
| Soil 1 | Penicillium funiculosum | 99 | 100 | 0 | HM017065.1 |
| Soil 1 | Bipolaris sp. | 100 | 100 | 0 | KF553640.1 |
| Soil 1 | Colletotrichum boninense | 98 | 98 | 0 | JQ754138.1 |
| Soil 2 | Curvularia sp. | 100 | 100 | 0 | HG779068.1 |
| Soil 2 | Trichoderma sp. | 99 | 99 | 0 | AB373724.1 |
| Soil 2 | Penicillium funiculosum | 100 | 99 | 0 | HM017065.1 |
| Soil 2 | Bipolaris sp. | 100 | 100 | 0 | KF553640.1 |
| Soil 2 | Bjerkanderasp. | 98 | 100 | 0 | KF698751.1 |
| Soil 2 | Penicillium camemberti | 100 | 99 | 0 | NG_042320.1 |
| Soil 3 | Alternaria alternata | 99 | 100 | 0 | KP124545.1 |
| Soil 3 | Penicilliumsp. | 97 | 100 | 0 | HM367083.1 |
| Soil 3 | Penicillium sp. | 97 | 100 | 0 | HM367083.2 |
| Soil 3 | Bjerkandera adusta | 98 | 100 | 0 | AB733334.1 |
| Soil 3 | Curvularia lunata | 99 | 99 | 0 | KF590137.1 |
| Leaf litter 1 | Trichoderma sp. | 99 | 99 | 0 | AB373724.1 |
| Leaf litter 1 | Talaromyces sp. | 99 | 99 | 0 | AB808325.1 |
| Leaf litter 1 | Penicillium verruculosum | 98 | 100 | 0 | JX863916.1 |
| Leaf litter 1 | Fusarium sp. | 99 | 100 | 0 | KJ850949.1 |
| Leaf litter 2 | Trametes hirsuta | 99 | 99 | 0 | AB733344.1 |
| Leaf litter 2 | Hirsutella fusiforme | 100 | 99 | 0 | KJ524706.1 |
| Leaf litter 2 | Trichoderma sp. | 99 | 99 | 0 | AB373724.1 |
| Leaf litter 3 | Bipolaris sp. | 100 | 100 | 0 | KF553640.1 |
| Leaf litter 3 | Trichoderma sp. | 99 | 99 | 0 | AB373724.1 |
| Leaf litter 3 | Trichoderma sp. | 99 | 99 | 0 | AB373724.1 |
| Leaf litter 3 | Bjerkandera sp. | 98 | 100 | 0 | KF698751.1 |
| Leaf litter 3 | Penicillium sp. | 97 | 100 | 0 | HM367083.1 |
| Leaf litter 3 | Talaromyces funiculosus | 99 | 100 | 0 | KP784313.1 |
Higher cellulase activity was observed in Trichoderma sp. isolated from the soil in point 3 and Colletotrichum boninense isolated from the soil in point 1 (Table 2). All the strains were positive for at least one of the tests and eleven exhibited lipase activity in only one test, revealing that temperature can affect the lipase activity of these fungi (Bentubo & Gompertz, 2014; Naz & Jadhav, 2015). All the strains were tested for laccase production in mediums containing guaiacol (4 mM), but none produced this enzyme under the applied conditions (data not shown).
Table 2: Cellulase and lipase activitiesfrom fungi isolated from soil and leaflitter collected in Brazilian savanna
| Sample | Taxa | Cel. | Halo (cm) | Mean | EI* | Lip. 1 | Lip. 2 |
|---|---|---|---|---|---|---|---|
| S 1 | Pleosporales | + | 0.1 | 3.65 | 0.0274 | + | - |
| S 1 | Penicillium funiculosum | + | 0.1 | 0.85 | 0.1177 | + | + |
| S 1 | Bipolaris sp. | + | 0.35 | 4.65 | 0.0753 | + | + |
| S 1 | Colletotrichum boninense | + | 0.95 | 0.4 | 2.3750 | - | + |
| S 2 | Alternaria alternata | + | 0.4 | 3.6 | 0.1111 | + | + |
| S 2 | Penicillium sp. | + | 0.55 | 3.4 | 0.1618 | + | + |
| S 2 | Penicillium sp. | + | 0.1 | 3.35 | 0.0299 | + | + |
| S 2 | Bjerkandera adusta | + | 0.1 | 1.55 | 0.0645 | - | + |
| S 2 | Curvularia luneta | + | 0.2 | 4.65 | 0.0430 | + | + |
| S 2 | Bipolaris sp. | + | 0.5 | 4.65 | 0.1075 | + | - |
| S 3 | Penicillium sp. | + | 0.125 | 2.95 | 0.0424 | + | + |
| S 3 | Talaromyces funiculosus | + | 0.15 | 4 | 0.0375 | + | + |
| S 3 | Curvularia sp. | + | 0.5 | 4.55 | 0.1099 | + | + |
| S 3 | Trichoderma sp. | + | 0.5 | 0.35 | 1.4285 | + | - |
| S 3 | Penicillium funiculosum | - | 0 | 0 | 0 | + | + |
| LL 1 | Trichoderma sp. | + | 0.1 | 2.05 | 0.0488 | + | - |
| LL 1 | Talaromyces sp. | + | 0.2 | 0.3 | 0.6667 | + | - |
| LL 1 | Penicillium verruculosum | + | 0.1 | 0.5 | 0.2000 | + | - |
| LL 1 | Fusarium sp. | + | 0.1 | 0.95 | 0.1053 | + | + |
| LL 2 | Trichoderma sp. | + | 0.65 | 5.9 | 0.1102 | + | + |
| LL 2 | Trichoderma sp. | - | 0 | 0 | 0 | + | + |
| LL 2 | Bjerkandera sp. | + | 0.375 | 0.8 | 0.4688 | + | - |
| LL 3 | Bipolaris sp. | + | 0.45 | 3.9 | 0.1154 | + | - |
| LL 3 | Bjerkandea sp. | + | 0.25 | 0.7 | 0.3571 | + | + |
| LL 3 | Penicillium camemberti | + | 0.3 | 1.55 | 0.1936 | + | + |
| LL 3 | Trametes hirsuta | + | 0.1 | 0.65 | 0.1539 | + | - |
| LL 3 | Hirsutella fusiforme | + | 0.2 | 2.2 | 0.0909 | + | + |
| LL 3 | Trichoderma sp. | + | 0.1 | 1.15 | 0.0870 | + | - |
S = Soil, LL =Leaf litter, Cel. = Cellulase activity. Data are the mean values of two independent cultures. EI* is the enzymatic index (Hankin & Anagnostakis, 1975). Lip = Lipase. + indicates positive activity and -, negative, since halo was not observed in lipolytic activity assays. Lipase 1 was tested with phenol red while Lipase 2 was tested with Rhodamine B. The control strains for enzymatic activities were Pleurotus sp. for the cellulase test and Rhodotulura mucilaginosa for the lipase test. Bold indicates the strains with higher cellulase activity.
Discussion
Fungi of the Ascomycota phylum are usually abundant in soil samples and are generally widely distributed in soil types from arid to permafrost (Porras-Alfaro, Herrera, Natvig, Lipinski, & Sinsabaugh, 2011; Zhang et al., 2012). In Brazilian savanna soils, they correspond to at least 49 % of total fungi in riverbank forests and 61.2 % in soils, with percentages increasing when this soil is converted to pasture or to soybean culture (Castro et al., 2008). Ascomycota as well as Basidiomycota strains are influenced by dry and rainy seasons (Nogueira-Melo, Santos, & Gibertoni, 2014). The abundance of Basidiomycota, however, can be higher in gallery forest soil during the dry season, when organic matter increases (Castro et al., 2016). In the sampling in the present study, carried out at the beginning of the rainy season, Ascomycota were more abundant. In Brazilian savanna soils, the precipitation regime appears to be the most important factor for determining fungal diversity (Castro et al., 2016). Other factors, however, may influence the distribution of fungi, such as aluminum level, acidity, nutrient and water availability, which can interfere with savanna vegetation physiology (Oliveira-Filho & Ratter, 2002).
Respect to the fungi isolated from leaf litter samples, the Basidiomycota phylum is more common at the end of decomposition (Frankland, 1998), as the decay of lignin compounds form aromatic amines or phenols, which are generally degraded by peroxidases secreted by basidiomycetes known as “white-rot fungi” (Larrondo, Salas, Melo, Vicuña, & Cullen, 2003; Pozdnyakova, 2012). Only two Basidiomycota were isolated from leaf litter, one from point 2 (Trametes hirsuta) and one from point 3 (Bjerkandera sp.). The remaining 12 isolates belonged to the Ascomycota genera Fusarium, Penicillium, Trichoderma, Talaromyces, Bipolaris and Hirsutella. It is interesting to note that only four genera (Penicillium, Trichoderma, Bipolaris and Bjerkandera) occurred in the leaf litter and soil, indicating the specificity of some fungi. The cellulolytic fungi community changes over time during the decomposition of leaf litter (Frankland, 1998). The prevalence of Ascomycota in the samples of the present study might be explained by the fact that the leaf litter sampled was in the early stage of decomposition. It has been observed that Ascomycota has a higher relative abundance in temperate forests during the early stages while Basidiomycota increases in the latter stages (Vorísková & Baldrian, 2013).
Ascomycota phyla, such as the Penicillium and Trichoderma genera frequently isolated in the present study, are not as important in the degradation of lignin as Basidiomycota, although they can produce cellulolytic enzymes and chitinase, being more generalist and abundant than Basidiomycota (Baldrian et al., 2011). In a survey of the cellulolytic activity of strains isolated from different materials, the majority of strains with this metabolism were isolated from soil samples (Damaso et al., 2012), confirming soil as an important source of fungi for biotechnological studies. The IE showed by the Colletotrichum boninense strain isolated in this work was similar to that presented by the control strain Pleurotus sp. (2,2727), which indicates C. boninense as a good candidate for optimization aiming at cellulase production. It is possible that the conditions of the present study lacked an inducer for laccase production, as many fungi only express laccases in the presence of an inducer or under stressful conditions, such as low nitrogen availability or the presence of toxic substances and antibiotics (Martínez et al., 2005). Lipolytic activity was assessed under two different temperatures. Lipase secretion in endophytic fungi is higher under alkaline pHs and absent under acid pHs (Maccheroni Jr., Araújo, & Azevedo, 2004), but the effect of temperature in lipase production has not yet been assessed. A strain identified as C. gloesporioides, isolated from Cerrado soil, has been previously characterized as an effective alkaline lipase producer (Colen, Junqueira, & Moraes-Santos, 2006), and the strain of the same genera in the present study demonstrated lipase activity in the assay carried out at under 30 °C and pH 7.
The enzymatic activity of the filamentous fungi identified in this work reveals their biotechnological potential and reinforces the importance of assessing and preserving environmental resources. Fungi diversity might be reduced in soils from which vegetation has been removed (Barbosa et al., 2016) and the Brazilian savanna, from which these fungi were isolated, has been subjected to a high degree of deforestation and land-use change. This might cause the loss of these strains before their commercial potential can be assessed (Barbosa et al., 2016). With the increasing demand for renewable combustibles, the search for cellulolytic and lignocellulolytic microorganisms with high enzymatic activity is mandatory. The discovery of new strains might provide economic feasibility to second-generation biofuels, those obtained from lignocellulosic wastes, which is an important alternative to biofuels obtained from food crops.












 uBio
uBio 

