Introduction
Animal feed systems in the tropics. Livestock systems in tropical conditions are affected by variations in pasture productivity due to seasonal rainfall distribution and climatic phenomena such as tropical storms and hurricanes (Alfaro & Quesada, 2010). The use of conserved forage (hay, haylage and silage) and agro-industrial by-products as alternative food sources can help maintain the productivity of the system and the welfare of livestock during periods of the year when the demand for dry matter exceeds the forage supply (Pulido et al., 2020). Preservation of forage as silage is limited by the moisture content of the forage used, the soluble carbohydrate content, the compaction of the material (particle size and oxygen availability), the use of additives and the natural population of microorganisms in the material or mixture at the beginning of the process (Ávila & Carvalho, 2019). Non-compliance with the minimum requirements for an adequate fermentation process can lead to deterioration of the material to be preserved (loss of dry matter) and the growth of microorganisms that can affect the health of livestock and humans related to the production system (Auerbach & Nadeau, 2020). In addition, losses during cultivation, harvesting, transfer and compaction of the material to be ensiled (Kim et al., 2020) increase production costs (Villalobos-Villalobos et al., 2015). Undesirable microorganisms such as Clostridium, fungi, molds, yeasts, enterobacteria, Salmonella, and Listeria monocytogenes, are associated with the production of secondary metabolites (as mycotoxins and toxins) (Ávila & Carvalho, 2019); in addition, these microorganisms promote deamination, decarboxylation, and oxidation/reduction processes that lower the nutritional value of the material and reduce voluntary consumption by livestock (Queiroz et al., 2018). In particular, L. monocytogenes causes the food-borne disease listeriosis, which is associated with septicemia, encephalitis, meningitis, meningoencephalitis, rhombencephalitis, abortions, fetal death, perinatal infections, and gastroenteritis in livestock (Dhama et al., 2015). L. monocytogenes can remain on dairy cattle farms for months, even when good hygienic practices are applied. Poor quality silage is the main source of infection by this pathogenic bacterium (Idland et al., 2022; Yoshida et al., 1998).
Additives can be used to improve the fermentation process and reduce material losses by stimulating (bacterial culture and carbohydrate sources) or inhibiting fermentation (acids and salts of weak acids), inhibiting aerobic spoilage (propionic acid), and improving nutrient supply (urea) (McDonald, 1981). The addition of lactic acid bacteria (LAB) increases the initial number of these microorganisms and prevents the growth of enterobacteria and Clostridium (McDonald, 1981) by promoting the transformation of soluble carbohydrates to lactic acid (LA), which stabilizes the fermentation process. Because of their probiotic characteristics, LAB consumed in silage reduce the production of enteric methane (CH4), a greenhouse gas produced during the digestive process in ruminants (Huyen et al., 2020).
Microbiota present in silage made from agroindustrial residuals have not been evaluated and characterized in Costa Rica. Therefore, the objective of this research was to describe the dynamics of the microbiota present in king grass and pineapple silage during the fermentation process using next generation sequencing (NGS). The potential protective effect of L. paracasei_6714 against L. monocytogenes was also evaluated in inoculated silages.
Materials and methods
Microbial strains and inoculum preparation: L. paracasei_6714, an isolate from pineapple peel silage (WingChing-Jones et al., 2021) with antagonistic activity against pathogens was used in this assay (Wu-Wu et al., 2021). A 48 h culture of L. paracasei_6714 on De Man, Rogosa, and Sharpe agar (MRS) culture medium (Thermo Scientific™ Oxoid™, MA, USA) was used to prepare a suspension of 1 x 109 CFU in 100 ml of deionized water. L. monocytogenes strain ATCC 19116 was grown for 48 h on Tryptone Soy Agar (ATS) medium (Merck, NJ, USA) and inoculum was prepared as described for LAB.
Silage treatments: Silage was made from king grass (Cenchrus purpureus (Schumach.) Morrone, syn. Pennisetum purpuphoides) harvested after 70 days of regrowth and pineapple peel at a ratio of 90:10 (w/w). A total of 2 ml was added to forage and pineapple peel mixture (90:10, w/w), placed in transparent bags with a 2 kg capacity and a thickness of 0.0063 mm. The air was extracted with a vacuum pump and bags were sealed with elastic tape. The samples were kept on shelves in a laboratory under controlled conditions for 30 days at 25 ± 2 °C. The inoculation and collection of samples treated with L. monocytogenes was performed in a type two biosafety chamber (Labconco. MO, USA).
Three inoculated silage treatments and a non-inoculated control (CK) were used. Treatments were inoculated with either L. paracasei_6714 (CKLA), L. monocytogenes (CKLi), or a mixture of L. paracasei_6714 and L. monocytogenes (1:1) (CKLL) in the concentrations described above. Eight replicates of each treatment were prepared for a total of 32 bagged silages. At days 15 and 30 after the start of the fermentation process, two bags for each treatment and the control were opened and the dynamics of the microbiota were determined.
Nutritional value of silage after fermentation: After the fermentation period, a one kg sample of ensiled material was taken from each treatment and the dry matter (DM), crude protein (CP) (AOAC, 1998) and in vitro dry matter digestibility (IVDMD) were determined using the methodology of Van-Soest and Robertson, (1985). The pH was measured using a potentiometer with a hydrogen electrode and ammoniacal nitrogen was measured as described by Tobía et al. (2004), using 20 g of ensiled material in 80 ml of distilled water. Organic acids were extracted by ultrasound-assisted aqueous extraction under acidic conditions using a 1 g dry sample following the methodology described in AOAC with some modifications (Leiva & Granados-Chinchilla, 2020).
Statistical analyses: An unrestricted randomized design was used to evaluate the nutritional value of each silage. An analysis of variance (ANOVA) was performed with the statistical program SAS (SAS Institute Inc., 2011). When significant differences were determined as a result of the main effects, Duncan's test (P < 0.05) was applied to separate treatments.
16S rRNA sequencing: Forage and pineapple peels were sampled prior to fermentation. After 15 and 30 days of fermentation, 10 g composite samples containing material from three depths within the bag (5, 20 and 30 cm) were collected from each silage treatment.
Samples were homogenized with H20dd and DNA was extracted using the protocol described by Birnboim and Doly, (1979). The DNA was run in 1 % agarose gels. Samples were stored at -20 °C until further analysis. The DNA samples were sent to Macrogen Inc. (Korea) for the construction of genomic libraries. The 16S rRNA was amplified using the V3-V4 hypervariable region with the primers Bakt_341F (5'-CCTACGGGGGNGGGGCWGCAG-3') and Bakt_805R (5'-GACTACHACHVGGGGGTATCTAATCC-3') (Klindworth et al., 2013). Sequencing was performed using the MiSeq platform (Illumina.Inc), which yielded 300 bp fragments.
Bioinformatic analysis: Raw sequence data obtained from the Illumina sequencing platform were processed using QIIME2 (version 2018.11) (Bolyen et al., 2019; Caporaso et al., 2010) and its plugins. The 'dada2′ plugin (Callahan et al., 2016) was applied to truncate reads (--p-trim-left 0, --p-trunc-len 260). The vsearch uchime_ref method was used to identify chimeric feature sequences. Mitochondria sequences were removed. Taxonomy of the Amplicon Sequence Variant (ASV) was assigned using the SILVA 16S/18S rDNA non-redundant reference dataset (SSURef 132 NR) (Quast et al., 2013) with the feature-classifier classify-sklearn. Taxonomic assignments of the remaining ASVs were manually checked by comparing them with sequences in the database using a combination of initial BLASTN-based searches and an extension of the EzbioCloud (Yoon et al., 2017) which stores 16S rRNA gene sequences of type strains of validly published names.
All of the ASVs (4 432) were considered for graphics construction and diversity analysis. Shannon (S), Tajima (d) and Fisher indices were determined. A shadeplot of the biotic data (after square-root transformation of the original data-sheet) was created through the matrix display of the wizard tool, where real data are shown with samples and variables arranged and clustered according to their similarity (Bray Curtis similarity for samples and Whitaker index of association for ASVs).
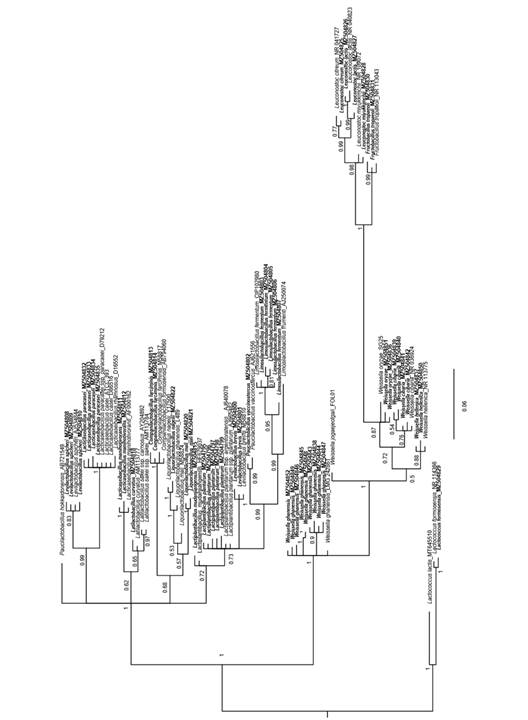
Phylogeny analysis: A phylogeny analysis was performed using all LAB isolates belonging to the family Lactobacillaceae that were present in the samples, considering the new classification proposed by Zheng et al. (2020) for the Lactobacillus group. The MUSCLE tool (Tamura et al., 2013) was used to align the sequences. The phylogeny was constructed by Bayesian analysis (Ronquist & Huelsenbeck, 2003) using 10 million generations, eight Markov chains and sampling every 1 000 generations. The sequences of Lactococcus formensis NR114366 and L. formensis (obtained in this research) were used as the outgroup.
Results
Silage quality - Nutritional parameters: Dry matter content and digestibility of silages did not differ significantly (P > 0.05) among treatments after 30 days of fermentation (Table 1). Crude protein content was highest in silage inoculated with L. monocytogenes alone and lowest in silage inoculated with both L. paracasei_6714 + L. monocytogenes. The CP content of silage inoculated with L. paracasei_6714 was not significantly different from that of the non-inoculated control.
Table 1 Nutritional and fermentative parameters of king grass and pineapple peel treatments after 30 days of storage.
| Treatments (%) | ||||
| Nutritional parameters | non-inoculated | L. paracasei_6714 | L. paracasei_6714 + L. monocytogenes | L. monocytogenes |
| Total dry matter | 20.17 (± 0.21) | 19.86 (± 0.49) | 19.63 (± 0.55) | 20.16 (± 0.51) |
| Crude protein | 10.78 (± 0.49)ab | 10.61 (± 0.12)ab | 10.01 (± 0.30)b | 11.21 (± 0.79)a |
| In vitro digestibility of dry matter | 64.97 (± 1.37) | 65.73 (± 1.06) | 63.90 (± 0.52) | 65.27 (± 1.06) |
| Fermentation parameters | ||||
| pH | 4.31 (± 0.12)b | 4.40 (± 0.06)ab | 4.43 (± 0.03)ab | 4.49 (± 0.04)a |
| Ammonia nitrogen /N-total | 4.90 (± 0.10)b | 5.33 (± 0.35)b | 30.40 (± 1.42)a | 26.83 (± 3.84)a |
| Lactic acid | 0.00 (± 0.00)b | 0.00 (± 0.00)b | 4.43 (± 0.36)a | 4.16 (± 0.61)a |
| Acetic acid | 7.94 (± 0.74)a | 6.30 (± 0.80)b | 0.50 (± 0.04)c | 0.49 (± 0.04)c |
| Propionic acid | 2.25 (± 0.25)a | 2.26 (± 0.34)a | 1.17 (± 0.17)b | 1.44 (± 0.26)b |
| Isobutyric acid | 0.39 (± 0.03)b | 0.35 (± 0.06)b | 0.58 (± 0.14)a | 0.63 (± 0.04)a |
| Butyric Acid | 5.14 (± 0.09)a | 3.10 (± 0.05)b | 0.11 (± 0.02)c | 0.15 (± 0.01)c |
| Total Acids | 15.72 (± 0.79)a | 12.01 (± 1.13)b | 6.69 (± 0.29)c | 6.87 (± 0.35)c |
Mean values (± standard deviation, n = 5). Different letters in the same row indicate significant differences of P < 0.05, according to Duncan's multiple range test.
Fermentation parameters: The pH was below 4.5 in all silage treatments. The pH in the treatment inoculated with L. monocytogenes was the highest and was statistically different from the control. Ammonia nitrogen values in treatments inoculated with L. monocytogenes were five to six times higher than in the control and in treatments inoculated with L. paracasei_6714 (P < 0.05). The LA and isobutyric acid concentrations were significantly higher in treatments inoculated with L. monocytogenes. In contrast the concentration of acetic acid (AA) was up to 14 times higher in treatments without L. monocytogenes. Total acid content in the non-inoculated control and the treatment with L. paracasei_6714 were 1.7 to 2.3 times higher than in the treatments inoculated with L. monocytogenes (Table 1).
Microbiota diversity of the silages: A total of 4 432 ASVs were obtained. Proteobacteria, Firmicutes, Bacterioidetes, Actinobacteria, Verrucomicrobia, Planctomycetes and Patescibacteria were the most abundant phyla. The relative abundance of each phylum varied depending on the material used and fermentation period. In the pineapple peel samples, the most abundant phyla were Firmicutes (80 %) and Proteobacteria (14 %), while the relative abundance of the other groups was less than 3 %. Proteobacteria (70 %) and Bacteroidetes (15 %) comprised up to 85 % of the phyla in king grass (Fig. 1). Despite the higher proportion of king grass in the silages and the important load of Proteobacteria, the abundance of Firmicutes was approximately 45 % at both sampling periods (15 and 30 days) in the treatments inoculated with L. paracasei_6714 and the non-inoculated control. In silage inoculated with L. monocytogenes, the abundance of Firmicutes was approximately 28 % at 30 days. The abundance of Proteobacteria remained near 40 % at both sampling times in all treatments (Fig. 1).
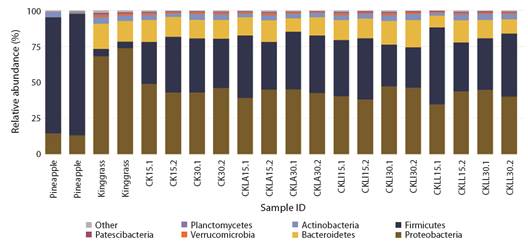
Fig. 1 Relative abundance of microbiota by phylum found in initial samples of pineapple and king grass and in inoculated and control silage treatments incubated for 15 and 30 days. CK: non-inoculated king-grass: pineapple peel (90:10) silage, CKLA: king grass: pineapple peel silage inoculated with L. paracasei_6714, CKLi: king grass: pineapple peel silage inoculated with L. monocytogenes (CKLi), CKLL: king-grass: pineapple peel silage inoculated with a mixture of L. paracasei_6714 and L. monocytogenes (1:1).
Structure of the bacterial community: The similarity of the communities at the species level using Bray-Curtis analysis was not greater than 50 %. In contrast, similarity at the phylum level was greater than 70 %. The main genera present in the pineapple peel were Weissella, Fructobacillus and Pseudomonas; species belonging to the Acetobacteraceae family were also present (Fig. 2). In king grass, several Pseudomonas species were found, including P. flevescens, P. fluorescens, and P. synxantha. Analysis of the bacterial community showed no treatment effect. After 15 and 30 days, genera not detected in the initial samples (pineapple and king grass) were present in the silages. These included Lactobacillus, Lacticaseibacillus, Levilactobacillus, Paucilactobacillus, Rhizobium and Leuctonostoc. L. monocytogenes was not found in any of the treatments inoculated with this bacterium or in the controls (Fig. 2).
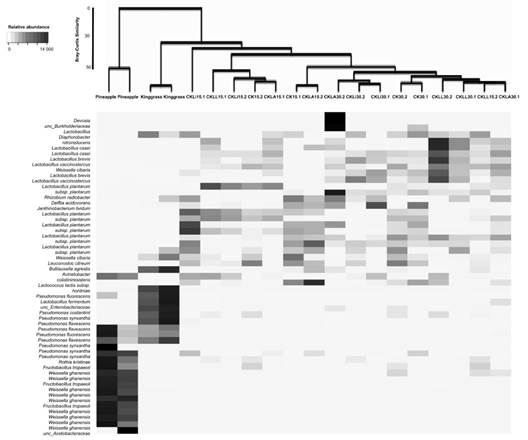
Fig. 2 Shadeplot of the main species present in the initial residues and treatments. CKLA: king grass: pineapple peel silage inoculated with L. paracasei_6714, CKLi: king grass: pineapple peel silage inoculated with L. monocytogenes (CKLi), CKLL: king grass: pineapple peel silage inoculated with a mixture of L. paracasei_6714 and L. monocytogenes (1:1). 15 and 30 days post inoculation (dpi) process.
Diversity analysis: Diversity values for the pineapple samples were lower than those of the king grass and fermented materials (Table 2). Average ASV values were highest in the control treatment (30 days post-inoculation (dpi)), followed by silage inoculated with L. monocytogenes (15 dpi). Shannon (S), Tajima (d) and Fisher indices of diversity were highest for the initial king grass material, followed by the control silage evaluated at 15 and 30 days (Table 2).
Table 2 Diversity indices determined for the Amplicon Sequence Variants (ASV) of the initial sample of pineapple, king grass and the silage treatments, 15 and 30 days post inoculation (dpi) process.
| Treatments | N | S | d | Fisher |
| pineapple | 42 871 (± 10 838) | 187 (± 46) | 19 (± 1,7) | 25 (± 6) |
| king grass | 71 485 (± 10 471) | 647 (± 114) | 58 (± 9) | 98 (± 18) |
| CK 15dpi | 84 702 (± 899) | 618 (± 7) | 54 (± 0,57) | 90 (± 1) |
| CK 30 dpi | 88 062 (± 10 989) | 622 (± 146) | 54 (± 12) | 90 (± 23) |
| CKLA15 dpi | 80 893 (± 2 970) | 575 (± 27) | 51 (± 2) | 83 (± 4) |
| CKLA 30 dpi | 68 798 (± 13 009) | 430 (± 148) | 38 (± 13) | 61 (± 22) |
| CKLI 15 dpi | 87 577 (± 8 062) | 609 (± 44) | 53 (± 3) | 88 (± 6) |
| CKLI 30 dpi | 76 109 (± 2 827) | 557 (± 72) | 49 (± 6) | 81 (± 12) |
| CKLL 15 dpi | 80 808 (± 1 734) | 553 (± 114) | 49 (± 10) | 80 (± 19) |
| CKLL 30 dpi | 77 280 (± 1 179) | 533 (± 35) | 47 (± 3) | 77 (± 6) |
N: number of ASV, S: Shannon, d: Tajima. Mean values (± standard deviation, n = 2). Abbreviations: CKLA: king grass: pineapple peel silage inoculated with L. paracasei_6714, CKLi: king grass: pineapple peel silage inoculated with L. monocytogenes (CKLi), CKLL: king grass: pineapple peel silage inoculated with a mixture of L. paracasei_6714 and L. monocytogenes (1:1).
Phylogenetic analysis of LAB species: A total of 53 ASVs were selected and deposited in GenBank with accession numbers MZ504795-MZ504853. A total of 33 sequences representing the main genera found in this research (marked in bold) were considered. The phylogenetic analysis confirmed the taxonomic classification of the ASV previously obtained using the NCBI and Ez Bio databases. There was a high level of support for most of the branches. ASVs corresponding to L. paracasei found in this research were affiliated with GenBank sequences of the same genus and with the close group of L. casei. These species are close to Levilactobacillus spicheri. Species more distant from L. paracasei in phylogeny were also identified and include Leuconostoc, Fructobacillus, and several species of Weissella (Fig. 3).
Discussion
King grass and pineapple residues are used for silage production in Costa Rica and other tropical countries (López-Herrera et al., 2019). In this research, physicochemical properties of these residues during the ensiling process were analyzed. Dry matter contains the main nutrients for animal nutrition and their loss during fermentation is a problem for animals (Ni et al., 2015; NRC 2001). The DM contents of LAB-inoculated materials in this work were similar to those reported by Santoso et al. (2015) and higher than those of Zi et al. (2021). The differences may be associated with the age at harvest of the forage material. High dry matter content indicates a high concentration of nutrients in the ensiled material and therefore a greater contribution to the livestock diet.
In our study, inoculation with LAB did not affect the DM content or the in vitro digestibility of the preserved material. This may have been due to the natural presence of LAB in the pineapple and king grass, which served as inoculum in the silage process (López-Herrera et al., 2014). Other studies have reported that inoculation with LAB increased DM content in LAB-inoculated materials (da Silva et al., 2018), increased in vitro digestibility (Cao et al., 2011), or had no effect on DM content (Santoso et al., 2015; Zi et al., 2021) of LAB inoculated materials.
The higher CP content of the silages treated L. monocytogenes may be due to the high ammoniacal nitrogen (a product of the degradation of amino acids during the fermentation process), and low total organic acids (a product of a less intense fermentation process). The pH values in all the treatments were lower than 4.5. A pH lower than 4.5 can inhibit the growth of undesirable microorganisms such as spoilage bacteria, yeasts, and pathogens (Rahman et al., 2014; Thierry et al., 2011). The lower pH of the control silage is the result of higher acid production during fermentation (Contreras-Govea et al., 2013). Both treatments inoculated with L. monocytogenes had higher pH values than the treatments not inoculated with the pathogenic bacteria. This result could be due to the presence of six times more ammonia nitrogen, which generates a buffer effect that reduces acidification of the silage (Gutierrez et al., 2003).
The high AA production in the silage treated with L. paracasei_6714 and the control could be explained by a fermentation pathway that promoted the transformation of sugars to AA. A heterofermentative pathway during ensiling stimulates the production of AA from the available carbohydrates (McDonald, 1981). In other trials, inoculation with heterofermentative bacteria, such as L. paracasei (Driehuis et al., 2002; Kleinschmit & Kung, 2006) improved the aerobic stability of silages due to the production of AA, which also inhibits the growth of yeasts (Guan et al., 2020). Comparison of the fermentative parameters showed silages with characteristics of an adequate tropical fermentative process. Fermentation is a spontaneous and continuous process; changes in the composition of the microbiota occur at different times, especially at the beginning when changes in pH occur (Muyzer et al., 1993; Ni et al., 2015).
The dominant bacteria present in pineapple peel were members of the genera Weissella and Fructobacillus, especially W. ghanensis and F. trapeoli, and members of the family Acetobacteraceae. Bacteria of the Weissella spp. have an important function at the onset of fermentation (Ni et al., 2015). These bacteria utilize water-soluble carbohydrates and convert them to CO2, water, and heat. During fermentation of whole-crop wheat, LA-producing bacteria, such as Weissella, initiate lactate fermentation and create anaerobic conditions that are suitable for the development of LAB (Ni et al., 2015). Pseudomonas was present in the initial king grass material. The role of some genera, including Pseudomonas, in silage processes has not been studied in detail (Yang et al., 2019). The presence of Pseudomonas in silages is undesirable due to their potential to produce biogenic amines (Ren et al., 2018). In the present study and in other research, the abundance of Pseudomonas in silage decreased over time (He et al., 2020).
The strain L. paracasei_6714 (used in this work) inhibited L. monocytogenes growth (Wu-Wu et al., 2021). Adding LAB inoculum has been shown to improve the ensiling process, protect ensiled material against the growth and survival of undesirable bacteria (Queiroz et al., 2018), and ensure silage quality (Driehuis et al., 2002, Wang et al., 2006).
In this research, NGS was used to examine the microbiota present in the ensiling of tropical forages and agro-industry residues. The use of this technique to characterize these processes is a relatively recent technology (Keshri et al., 2018; Keshri et al., 2019; Muraro et al., 2021; Wang et al., 2022; Zi et al., 2021) that allows a better understanding of the complex dynamics of microbial populations in silages (Keshri et al., 2019).
Our results suggest that inoculation with L. paracasei_6714 and L. monocytogenes can reduce the diversity of bacterial populations in silage. Diversity decreased over time (15 vs. 30 days) in inoculated silages but not in the non-inoculated control. Similar studies have shown lower diversity in silage treated with a lactobacillus compared with untreated silage (Duniere et al., 2017; Keshri et al., 2018; Romero et al., 2017; Wang et al., 2016). The decrease in diversity is attributed to the high concentration of the added inoculum and the inhibitory effect of the inoculated species on the growth and multiplication of other microorganisms (Keshri et al., 2018).
The presence of LAB during the fermentation process is important for the control of pathogenic microorganisms. When oxygen is consumed at the beginning of fermentation, anaerobic bacteria such as Lactobacillus grow and decrease the pH of the silage during early stages of fermentation (Wang et al., 2006). This could explain the presence of these microorganisms in the inoculated and control silages evaluated at 15 dpi. Most of the species found in this study have been previously reported in silages used for animal feed. For example, L. sakei was the dominant species in brachiaria grass (Brachiaria decumbes) during the first 10 days, while L. plantarum was present at all stages of storage and dominated in silage at 30 and 45 days (Santos et al., 2013). In other studies, L. brevis and L. buchneri predominated in maize grains stored for a prolonged period (Carvalho et al., 2017), L. paracasei produces a high concentration of AA that inhibits the growth of molds, and yeasts and helps to improve the aerobic stability of silages (Blajman et al., 2018).
Although two treatments were inoculated with L. monocytogenes, this microorganism was not detected in the sequencing analyses. Listeria has been reported in more than 50 % of fecal samples from animals without symptoms of listeriosis (Nightingale et al., 2004). This is attributed mainly to the ingestion of contaminated silage with high loads of this pathogen, although other causes have been reported (Meng & Doyle, 1997).
Silage of pineapple residues and king grass forage are used for animal feed in Costa Rica and other countries in the tropics (Araya & Boschini, 2005). To the best of our knowledge, this is the first study to evaluate the potential of LAB isolated in Costa Rica from agroindustrial pineapple silages to inhibit the growth of pathogenic microorganisms. The absence of L. monocytogenes suggests that growth of opportunistic pathogenic bacteria such as Listeria could be prevented with appropriate management of the silage process in the field. The risk of contamination is likely to increase when silages are exposed to air for feeding; multiplication of undesirable microorganisms may occur, and nutritional values may decrease (Driehuis & Oude-Elferink, 2000).
Proper management of silage processing can reduce and/or eliminate pathogenic bacteria. Inoculation with L. paracasei_6714 did not improve the physicochemical characteristics or the associated microbiota of pineapple peel and king grass silage.
This study highlights the importance of nutritional and microbiological characterization of residues used for animal feed. In addition, the correct preparation of silages can help control pathogenic microorganisms in the field. Future research should consider other storage conditions, such as temperature and humidity, and evaluate potential protective effects of inoculated LAB against mycotoxin-producing fungi.
Ethical statement: the authors declare that they agree with this publication and that each made significant contributions; that there is no conflict of interest of any kind; and that we followed all pertinent ethical and legal procedures and requirements. All financial sources are fully and clearly stated in the acknowledgment section. A signed document has been filed in the journal archives.












 uBio
uBio