Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.56 n.3 San José Sep. 2008
The impact of local extinction on genetic structure of wild populations of lima beans (Phaseolus lunatus) in the Central Valley of Costa Rica: consequences for the conservation of plant genetic resources
Daniel Barrantes1, Gabriel Macaya2, Luigi Guarino3, Jean Pierre Baudoin4 & Oscar J. Rocha1,5
1. Escuela de Biología, Universidad de Costa Rica, Ciudad Universitaria Rodrigo Facio, San Pedro de Montes de Oca, San José, Costa Rica.
2. Centro de Investigación en Biología Celular y Molecular, Universidad de Costa Rica, Ciudad Universitaria Rodrigo Facio, San Pedro de Montes de Oca, San José, Costa Rica.
3. International Plant Genetic Resources Institute, Regional Office for the Americas, IPGRI c/o CIAT, A.A. 6713. Cali, Colombia.
4. Department of Tropical Crop Husbandry and Horticulture, Gembloux Agricultural University, 2 Passage des Déportés, B. 5030 Gembloux, Belgium.
5. Department of Biological Sciences, Kent State University, Kent, Ohio, 44242, USA; orocha@kent.edu
Abstract: Plant populations may experience local extinction and at the same time new populations may appear in nearby suitable locations. Species may also colonize the same site on multiple occasions. Here, we examined the impact of local extinction and recolonization on the genetic structure of wild populations of lima beans (Phaseolus lunatus) in the Central valley of Costa Rica. We compared genetic diversity from the samples taken from the populations before and after extinction at 13 locations using microsatellite markers. Locations were classified according to the occurrence of extinction episodes during the previous five years into three groups: 1) populations that experienced extinction for more than one year, and were later recolonized (recolonized), 2) populations that did not experience local extinction (control), and 3) populations that did not experience local extinction during the study, but were cut to experimentally simulate extinction (experimental). Our data did not show a clear tendency in variation in allele frequencies, expected heterozygosity, and effective number of alleles within and between groups of populations. However, we found that the level of genetic differentiation between samples collected at different times at the same location was different in the three groups of populations. Recolonized locations showed the highest level of genetic differentiation (mean Fst= 0.2769), followed by control locations (mean Fst= 0.0576) and experimental locations (mean Fst= 0.0189). Similar findings were observed for Neis genetic distance between samples (di,j= 0.1786, 0.0400, and 0.0037, respectively). Our results indicate that genetic change in lima beans depends on the duration and frequency of local extinction episodes. These findings also showed that control populations are not in equilibrium. Implications of these results for the establishment of conservation strategies of genetic resources of lima beans are discussed. Rev. Biol. Trop. 56 (3): 1023-1041. Epub 2008 September 30.
Keywords: genetic diversity, metapopulation, genetic differentiation, soil seed bank, in situ conservation, lima beans, Phaseolus lunatus, local extinction, recolonization, genetic resources.
It is well known that wild populations of plants may go extinct on a local scale, and new populations may appear at other suitable nearby sites (Levins 1969, Hanski 1991, Hanski 1991, Hanski and Gilpin 1991, Husband and Barrett 1996, Álvarez-Buylla et al. 1996, Sork et al. 1999). Over the course of time, a species may colonize the same site on multiple occasions, as ephemeral habitats disappear and form again periodically (Poschlod 1996, Gillman 1997, Menges et al. 1998, Rocha et al. 2002). These phenomena cause popula tions to fluctuate in size and to change in their spatial distribution (Menges 1990, Husband and Barrett 1996, Álvarez-Buylla et al. 1996). The term metapopulation has been coined to describe circumstances where individuals of a target species are found in groups or assemblies of dynamic, interacting populations that need to be studied as a whole in order to obtain an accurate portrayal of the species. Metapopulation models have the advantage of recognizing that local populations are dynamic (Harrison and Quinn 1989, Silvertown 1991, Olivieri 1990, Hanski 1991, Sork et al. 1999).
Metapopulation dynamics also affects genetic structure and population diversity and differentiation (Slatkin 1977, 1987, Wade and Mc Cauley 1988, Mc Cauley 1993, Gliddon and Goudet 1994, Giles and Goudet 1997, Pannell and Charlesworth 2000). Slatkin (1977) pointed out that there are two factors determining the effects of local extinction and recolonization on genetic differentiation: 1) a sampling process of individuals from the previous population to form the propagules that recolonize the site, and 2) the dispersal of propagules between populations. In general, the direction and magnitude of the effects of an extinction episode depend on the role that population size, genetic drift, founder effect, and pollen movement and migration play (Slatkin 1977, 1987, Wade and Mc Cauley 1988, Hamrick and Godt 1989, Hanson 1991, Mc Cauley 1993, Ellstrand and Elam 1993, Álvarez-Buylla et al. 1996, Husband and Barrett 1996, Hanski and Gilpin 1997).
Gene flow (pollen movement and seed migration) is an important factor determining genetic structure at the landscape level. Metapopulation structure may break down under high disturbance rates (valverde and Silverton 1997, Husband and Barret 1996, Ouborg et al. 1999). Habitat fragmentation and patch isolation may lessen the probability of recolonization events (Giles and Goudet 1997, Gillman 1997, Ouborg et al. 1999). Thus, isolation by distance may result in a disruption of the structure of the metapopulation (Giles and Goudet 1997, Gillman 1997). On the other hand, some authors have shown that gene flow appears to be high for tropical plants even in severely disturbed landscapes (Nason et al. 1997, White and Boshier 2000, Rocha and Aguilar 2001, Céspedes et al. 2003).
Metapopulation studies provide a new approach to study the ecology and genetics of populations at the landscape level that is of special interest for conservation biology (Gliddon and Goudet 1994, Hanski 1999). Human activity creates metapopulation scenarios, e.g. intensive land use, changes in agricultural practices, and urban development result in fragmentation of the landscape and temporal degradation of suitable habitats (Rocha et al. 1997, Rocha et al. 2002, Barrantes 2003). However, more information is needed about the impacts of human activities on dispersal, as it plays a key role in maintaining the integrity of metapopulations.
This study was part of a project which systematically gathered detailed ecogeographic, demographic, phenological and genetic data, and used these to develop and implement an effective, sustainable conservation strategy for a wild crop relative, the wild lima bean (Baudoin et al. 2004). Wild and weedy relatives of cultigens are an extremely important source of genes for crop improvement, but these crucial genetic resources are coming under increasing pressure the world over, as wild habitats are altered and lost. Previous studies have revealed the occurrence of multiple extinction and recolonization events in lima beans (Rocha et al. 2002, Barrantes 2003), but their consequences on genetic diversity have not been studied in detail.
Here, we examine the impact of local extinction on the levels of genetic diversity and differentiation of wild populations of lima beans. We sampled locations where wild lima beans disappeared for more than one year and later recolonized the site. We compared these data with those from control populations that did not experience extinction events during the same period of time. We also conducted an experimental removal of all plants in selected locations and examined its impact on the genetic diversity of the population that recolonized the site.
Materials and methods
Study area: This study was conducted in the Central valley of Costa Rica. This region is located in the geographic center of the country and comprises two intermontane valleys separated by the low Ochomogo pass at the continental divide: the Central valley of the Pacific slope and the much smaller valle del Guarco on the Caribbean side. However, most of this study was conducted in the upper valley and watershed of the Río Grande de Tárcoles (latitude range 9º 54 N-10º 07 N, longitude range 83º 50 W-84º 28 W). We also included in this study a portion of the valle del Guarco and the small valley of Acosta. As a whole, the study area is approximately 2,100 km2.
In early November 1992, we started a large-scale survey of populations of Phaseolus lunatus L. in the study area. When we surveyed a given area, we examined all roads and trails looking for plants. We also interviewed local farmers in order to find out whether the species was present in the area or not, and asked for permission to visit their farms. Where P. lunatus was found, we defined as one population any group of plants separated (regardless of its size) by at least 500 meters from other plants. For each population, we collected a voucher specimen, fruit and seed samples, and described the site with parameters such as size of the population, topography, associated vegetation and geographical coordinates using a Garmin 48 GPS unit. Similar surveys were conducted again in 1993, 1994 and 1999.
Previous studies revealed the occurrence of multiple extinction and recolonization events among wild populations of lima beans (Rocha et al. 2002, Barrantes 2003). Monthly monitoring of 106 populations for six year (from November 1994 to December 2000) showed that 39 populations experienced local extinction that lasted for more than one year. Only sixteen of these locations were recolonized (41%) in later years. The data also that only 28 populations produced seeds every year, indicating that a sizable fraction of the populations do not contribute to genetic flow among populations. However, many populations, including
those that produced seeds every year, often experienced anthropogenic disturbances that diminished their size (Barrantes 2003).
Model species: The wild lima bean has a climber habit and it is considered a short-lived perennial species. Typically, it grows during the rainy season; while in the dry season it loses its leaves and remains dormant. Dispersion of seeds takes place by means of the explosive opening of the pods (Degreef et al. 1997, Degreef 1998, Zoro Bi 1999). An important characteristic of P. lunatus is its ability to maintain reserves of seeds in the soil. These seeds can remain viable for up to three years after being dispersed (Degreef et al. 1997, Degreef 1998). The dormancy of seeds is induced during the dry season in response to dry conditions. Germination is induced by the increase in soil humidity at the beginning of the rainy season (Degreef et al. 1997, Degreef 1998, Degreef et al. 2002).
Lima bean flowers are very similar to those of the common bean (P. vulgaris). They present themselves in pseudo-clusters and they require the visit of insect pollinators to transfer the pollen successfully from the anthers to the stigma. The species is self-compatible and inbreeding is very common; however, the outcrossing rate can vary significantly within and between populations (Maquet et al. 1996, 1997, Zoro Bi 1999, 2003, 2005).
Effects of local extinction: In order to determine the effect of local extinction and recolonization on genetic diversity at a given location, the genetic structure of 12 populations was studied using microsatellite markers. The selected locations were classified into three groups according to the occurrence of extinction episodes during the previous five years (Table 1) and the availability of seeds from previous years in our seed bank collection. In the first group were sites where the population experienced local extinction for more than one year, and were later recolonized (recolonization). In the second group were populations that did not experience local extinction during the study period, but were cut to experimentally simulate an extinction episode (experimental). In the last group were populations that did not experience local extinction during the study period, and served as control (control). The selected populations were distributed throughout the Central valley, and their geographical locations are shown in Table 1. For each location, genetic diversity was compared between the samples taken from the populations present at two different times. At least twenty-five individuals were randomly chosen from the population present at each collecting date.
Four locations were included in the recolonization group. The populations present in these sites suffered at least one period of local extinction and recolonization. In addition, these locations were selected because there were seeds, collected prior to the extinction episode, stored in a seed collection maintained as part of this project. This group included sites Her2, E17, E29 and E38. These populations differed in the number of extinction episodes that they expe rienced and the number of years in which they produced seeds. Location E17 experienced three extinction episodes during our study, and populations E38, HER2, and E29 experienced only one. However, location E38 and HER2 were recolonized two years after extinction, while location H29 was colonized after one year.
Five locations were included in the second group (experimental). These locations showed similar conditions to those of the control group, i.e. no history of local extinction. A sample of 25 individuals was collected from each of these populations, and then all lima bean plants pres ent were cut. All experimental manipulations were conducted between May 2000 and April 2001, i.e. before the time of fruit maturation and seed dispersal. All sites were recolonized the following rainy season. In order to compare the genetic diversity of the populations present before and after the experimental manipulation, leaf tissue was collected from 25 individuals from each newly established population. Two of these populations (E104 and E25) were exposed to a partial disturbance, letting a small patch of isolated individuals complete its reproduction period and disperse seeds. In addition, it is important to point out that population E25 was selected as control population (samples from 1995 and 2000) and experimental population (samples from 2000 and 2001). These transient extinction events are frequently observed in lima bean populations throughout the Central valley.
The last group of locations included sites that had not experienced local extinction since November of 1994 (control). In addition, these populations were successful in producing seeds every year. In this case, genetic diversity was compared between a sample of seeds collected in the same location in 1995, and a sample of the individuals present in May 2001.
DNA extraction: Total genomic DNA was extracted from leaves of lima beans using the protocol developed by Lodhi et al. (1994), with some modifications based on the protocol proposed by Centro de Investigación en Agricultura Tropical (CIAT) for DNA extraction from common bean (González et al. 1995). Approximately 250 mg of leaf tissue was ground in liquid nitrogen with a pestle in a 1.5 ml Eppendorf tube. The ground tissue was mixed with 600 (l of 0.1 M Tris-HCl pH 8.0, 1.0 M NaCl, 2% CTAB, 2% PvP, and 1 % ( mercatoethanol extraction buffer. The mix was incubated at 60 (C for 40 minutes with agitation every 10 minutes. The samples were then centrifuged for 10 minutes at 12,000 g at 10 (C. The supernnatant solution was placed in another tube, and one volume of chloroformoctanol (24:1) was added, mixed gently, and centrifuged for 10 minutes. The supernatant was transferred to another tube where 1/10 volume of 3M NaAC pH 5.2 and 0.6 volume of ice-cold isopropanol was added. The mixture was kept at 5 (C for 10 to 20 hours. DNA was precipitated with 70% ethanol, air dried, and later resuspended in 100 (l of 1.0 M TE buffer.
Genetic analysis: Five microsatellites marker genes were used to determine genetic diversity. Primers for these loci were developed by Yu et al. (2000) for common bean (P. vulgaris). Table 2 describes the forward and reverse primers for each microsatellite locus, and the conditions for the polymerase chain reactions (PCR). PCR reactions were conducted using an air heated Rapid Cycler (Idaho Tecnology), and the reaction products were visualized on 40 cm long polyacrilamide gels using silver staining (Promega). The different alleles for each locus were identified according to their molecular weight, where the most anodal allele was arbitrarily identified as "1", and the remaining alleles numbered sequentially.
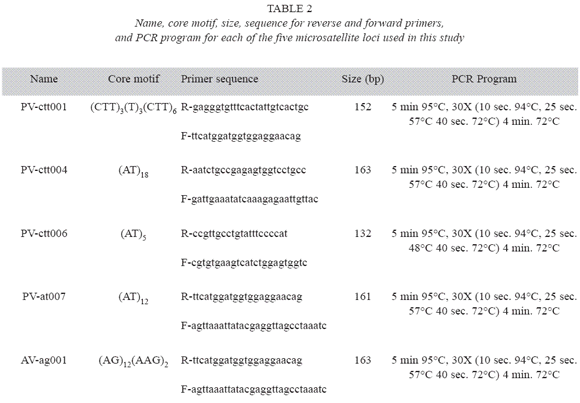
Data Analysis: Genetic diversity in each sample was quantified in terms of allele frequencies, number of alleles per locus (A), effective number of alleles per locus (AE), observed heterozygosity (HO), and Neis expected heterozygosity (HE) (Nei 1973) averaged over all loci. The significance of allele frequency differences between locations was assessed using the exact test (Weir 1996). The effective number of alleles was estimated as the reciprocal of the homozygosity (Hartl and Clark 1989). In addition, genetic differentiation was determined using the infinite allele model Fst (Weir and Cockerham 1984). The degree of relatedness between locations, based on Neis genetic distances, was represented in a tree using UPGMA. Bootstrap sampling of loci tested the robustness of clusters of the tree, using 103 simulations. These analyses were conducted using the program POPGENE 1.31 (Yeh et al. 1999).
Results
Genetic diversity: In order to study the genetic diversity of the three groups of populations we determined the allelic frequencies and the number of alleles for three microsatellite loci. We only found one allele in two of the loci that we examined; namely, loci PV-AT007 and VA-AG001. Table 3 shows the allelic frequencies for the three polymorphic loci for each population, sampling year, and population group. The data revealed that the populations in the first group (recolonized) tended to experience an increase in the frequency of the most common allele at locus PV-AT004. For locus PV-ctt001, the trend in allele frequencies is not clear. In population E29, the most common allele at that locus became fixed, and the least frequent allele disappeared from the population. However, in the other three populations the most common alleles before the extinction episode experienced a reduction in frequency. Our data also revealed significant heterogeneity in allele frequencies observed at locus Pv-ctt001 between samples taken at different times (1994-1995 vs. 2000-2001) in the same location for all populations (Table 4).
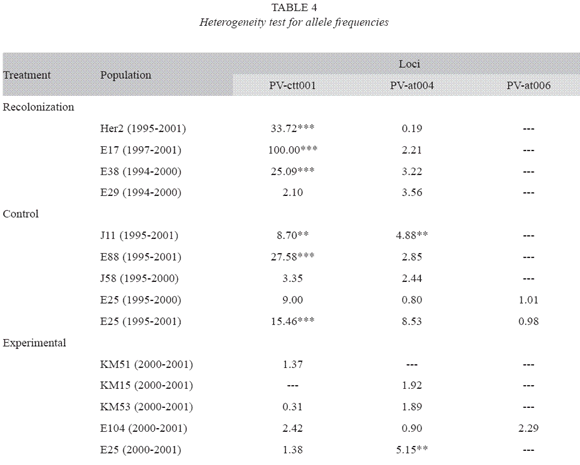
We did not find a clear trend in the variation in allele frequencies in the control population (Table 3). The most common allele at locus PV-ctt001 only experienced an increase in frequency in two populations (J58 and E25), while the same only happened in one population (E25) at locus PV-at004. Overall, these findings revealed significant changes in allele frequencies over the study period in the control populations. We found significant heterogeneity in allele frequencies between samples taken at different times from the same location for all populations at locus PV-ctt001 and for two populations at locus PV-at004 (Table 4).
There were also changes in allele frequencies in the samples taken from locations subjected to experimental manipulation (Table 3). For example, our data revealed that in three of the five locations the allele with the lowest frequency disappeared at locus PV-at004. There was only a minor change in allele frequencies at the same locus for the other population. However, the changes in allele frequencies at locus PV-ctt001 were more variable. We did not find major changes in allele frequencies in two populations (Km15 and Km53). We observed the appearance of a new allele in one population (E104), an increase in the frequency of the most abundant allele in one population (Km51). We also recorded the appearance of a new allele at locus Pv-at006 in one population (E104).
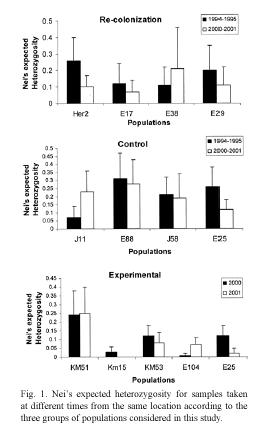
Genetic diversity, as indicated by Neis expected heterozygosity, tended to be affected by extinction (Fig. 1). Three recolonized locations showed a reduction in expected heterozygosity and one showed an increase. In contrast, there was no clear tendency in the change in heterozygosity in the populations in control locations. Similarly, there was no clear tendency among populations in experimental locations.
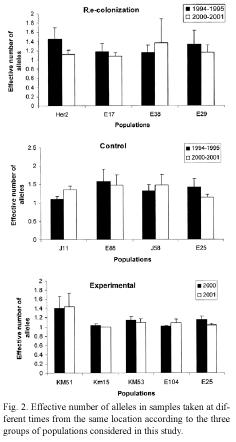
Our data also revealed changes in the effective number of alleles in the three groups of populations (Fig. 2). For example, three of the four locations in the recolonization group showed a reduction in the effective number of (E25), and a decrease in the frequency of the alleles. In contrast, one location (E38) showed most abundant allele in another population an increase in the effective number of alleles. Variation in the effective number of alleles was also observed in locations in the control group, where effective number of alleles increased in two populations but decreased in the other two. Only one population (E25) showed an important change in the effective number of alleles among the experimental populations (from 1.15 to 1.03).
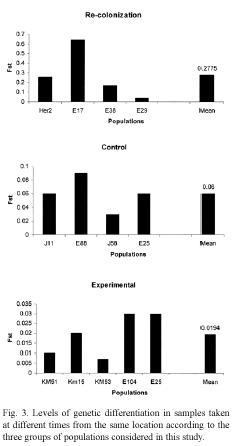
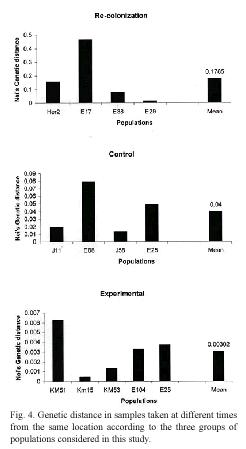
Genetic differentiation: The impact of local extinction on genetic diversity was also investigated by examining the genetic distance and the level of population differentiation in samples collected at different times at the same location. The data indicate that genetic differentiation and genetic distance between populations sampled in each location are affected by the occurrence of extinction events (Fig. 3). In locations where lima bean populations experienced long term extinction and subsequent recolonization, the level of genetic differentiation (Fst) ranged from 0.0407 to 0.6371 (mean Fst = 0.2769). In contrast, in control locations, genetic differentiation between the samples collected in 1995 and that of 2000 or 2001 ranged between 0.0263-0.0860 (mean Fst = 0.0576). Similar findings were observed for genetic distance between populations: for recolonized locations genetic distances were, on average, larger than that of control populations (Nei´s genetic distance, 0.1786 and 0.0400 respectively).
The populations subjected to experimental manipulation showed the lowest level of genetic differentiation (mean Fst = 0.0189), and also the lowest genetic distance (mean Nei´s genetic distance = 0.0030) between plants present before and after the transient extinction (Fig. 4). These findings indicate that populations of lima beans may experience short anthropogenic extinction and still preserve their genetic diversity. These populations successfully produced seeds every year, except for the year when they were cut. In these locations, a new population of lima beans was easily reestablished after the experimental manipulation.
Discussion
We evaluated the changes in genetic structure after the occurrence of local disturbances that caused population extinction in wild populations of P. lunatus in Central valley of Costa Rica. Demographic information revealed the occurrence of frequent events of local extinction and recolonization, suggesting a metapopulation structure for this species (Degreef et al. 1997, Rocha et al. 1997, Degreef 1998, Rocha et al. 2002, Barrantes 2003). Our results revealed a complex dynamic in the genetic structure of the populations considered in this study, which includes changes in the allele frequencies, loss and gain of alleles, and changes in the levels of population differentiation between samples collected at different times at the same location. Overall, the magnitude of these changes is, on average, accelerated by prolonged local extinction.
Genetic diversity: The level of genetic variation within populations has received considerable attention because it is considered an indicator of its evolutionary potential to respond to environmental changes (Frankel and Soulé 1981, Hamrick and Godt 1989, Heywood 1991, Newman and Pilson 1997, Hanski 1999, Cruzan 2001). It has been argued that losses in viability are caused by reduction in the number of alleles due to increases in endogamy and genetic drift (Frankel and Soulé 1981, Ellstrand and Elam 1993). Such losses may be the result of, among other causes, the fixation of deleterious alleles or the loss of heterozygous advantage (Ellstrand and Elam 1993, Newman and Pilson 1997, Cruzan 2001). Our results indicate that the level of disturbance, as well as time elapsed between sampling, influenced the magnitude of the changes in the allelic frequencies and the number of alleles present in populations. Local extinction increased the variation in allelic frequencies, favoring allele loss in sites where lima beans experienced extinction and recolonization. In contrast, experimental populations that only experienced transit extinction tended to preserve their alleles. In addition, these populations also experienced small changes in allele frequencies. Overall, variation in allelic frequencies was larger among populations in recolonized sites than among the populations in the other two groups.
Several factors might explain the observed changes in allele frequencies. The number and the duration of the extinction episodes can explain, at least in part, some of these changes. For example, the number of extinction episodes was variable among populations in recolonized sites. One population (E17) experienced two extinction episodes during our study, while populations E38, HER2, and E29 experienced only one. As predicted, genetic change was larger in populations that experienced multiple extinction episodes. In addition, the duration of the extinction episodes in location E17 lasted for more than 15 months each. In locations E38 and E29 the only extinction episode lasted for nearly 22 months, and in location E29 it only lasted 12 months.
It has been argued that local extinction results in population bottlenecks and elevated inbreeding during the recolonization of the site (Pannell and Dorken 2006). In this study, population E17, from the recolonization group, not only experienced two extinction episodes, but also showed the smallest mean population size during the study period. It is expected that under such conditions inbreeding should be high. Among the other three populations in this group, population E29 showed the largest mean population size, and shows the lowest Fst. These findings suggest that multiple extinctions lead to more severe bottlenecks, hence resulting in more differentiation between samples taken form the same location. Moreover, the likelihood of extinction is also related to population size, as small populations are more likely to disappear (Rocha et al. 2002, Barrantes 2003), which suggests that such populations may be more susceptible to genetic losses.
Extinction episodes are often related to failure of reproduction, and hence facilitate changes in genetic diversity (Degreff 1998, Rocha et al. 2002). Absence of seed production has a negative effect on the size and the genetic diversity of the soil seed bank (Kalisz et al. 1997, Degreef et al. 2003). Plants in population E17 failed to produce seeds during the study period, and this population experienced the greatest genetic change during this study. It is possible to argue that such failure to reproduce significantly affects the size of the soil seed bank of lima beans, hence the population that recolonized this site was probably not a random sample of the population previously present. Studies of the dynamics of the seed bank revealed that nearly 98% of the seed in the soil germinate or die within three years after dispersal. This finding supports the notion that the seed bank may be depleted in some of these populations (Degreef et al. 2002). On the other hand, seeds were produced almost every year in location E29, and the integrity of the seed bank may explain the low level of genetic differentiation between samples taken at different times.
Variation in allele frequencies was also observed in control populations. These sites had large populations, and were not expected to suffer the negative effects of genetic drift; however, they all experienced significant genetic change. In two populations (J11 and E88), the most abundant allele experienced a reduction in frequency at all polymorphic loci. In one population (E25), the most abundant allele experienced an increase in frequency at all polymorphic loci. In another population (J58), the most abundant allele experienced an increase in frequency at one polymorphic locus and a reduction at another. These findings strongly suggest that these populations are not in equilibrium. These changes might result from the arrival of genes from adjacent populations, as the level of genetic differentiation between populations is high (Maquet et al. 1997, Zoro Bi et al. 2003), or from complex dynamic processes resulting from within populations structure (Zoro bi et al. 1997) and/or changes in seed dispersal within populations (Hardy et al. 1997).
Experimental populations underwent the smallest changes in allele frequency among the loci examined. Most of these changes were not significant. We suggest that this limited change illustrates the role of a healthy soil seed bank in the conservation of genetic diversity. Our findings suggest that a constantly renewed soil seed bank maintained a representative sample of the allele frequencies found in the population. The levels of genetic change observed in these populations may result from sampling error, or internal dynamics driving these populations to equilibrium.
Changes in expected heterozygosity can also serve as an indicator of the impact of local extinction on genetic diversity. It has been proposed that the low allelic richness and heterozygote deficiency observed in wild populations of lima beans could be attributed to frequent population bottlenecks, founder effect, steady selfing, assortative mating, biparental imbreeding, selection favoring homozygote individuals and Wahlund effect (Rocha et al. 1997, Zoro Bi 1999, Zoro Bi et al. 1997, 2003, Rocha et al. 2002). The majority of the populations considered in this study showed a reduction in heterozygosity. On average, the locations that suffered local extinction experienced greater reduction in heterozygosity. This contrasts with populations in control locations, which exhibited both increases and reductions in heterozygosity. Moreover, despite having larger initial heterozygosity, control location exhibited, on average, smaller reductions in this parameter than recolonized locations. In addition, control populations were, in general, larger than populations in recolonized locations. Finally, experimental populations showed the smallest reduction in heterozygosity. In general, these findings support the notion that local extinction reduces genetic diversity, but our findings indicate that it is not the only factor involved.
Genetic differentiation: Several studies have examined the effect of extinction-colonization on genetic differentiation (Slatkin 1977, Wade and McCauley 1988, McCauley 1993, Barton and Whitlock 1997). These studies show that local extinction and recolonization can enhance or reduce genetic differentiation between populations as result of gene flow (Hanski 1999, Pannell and Charlesworth 2000, Pannell and Dorken 2006). Whitlock and McCauley (1990) proposed that the effect of extinction and recolonization depends on two key factors: the patterns of gene flow and the number of individuals establishing the new population. Overall, we found that genetic differentiation between populations increased with time. In 1994-95, the genetic differentiation was smaller than was observed in 2000-01 (F= 0.34 and Fst= 0.42, respectively). These values are similar to those reported by Zoro Bi et al. (2003) among 29 wild populations of lima beans using 23 isozyme loci. However, significant genetic differentiation between samples taken from each location over time was also observed in this study.
Overall, samples taken from recolonized populations in 1994-95 and 2000-01 revealed the highest level of differentiation and genetic distance, followed by control (1994-95 and 2000-01) and experimental populations (2000 and 2001). The levels of genetic differentiation that we found suggest that, in recolonized populations, there is limited gene flow between samples. This can result from the decay of the soil seed bank or from gene flow from adjacent populations. Moreover, our data also revealed that population E29, which only experienced one extinction episode, but where seeds were produced more frequently, showed lower differentiation than the other populations in this group. Meanwhile, population E17, which failed to produce seeds during this study, experienced the highest differentiation. Genetic differentiation between samples taken before and after experimental manipulation was also low, indicating that transient extinctions do not result in significant genetic change. This is especially true if lima beans have been producing seeds consistently prior to extinction.
A similar pattern is revealed when comparing genetic distances between samples. Our data revealed that the genetic distance between samples taken from the same location is larger at sites that experienced extinction episodes that lasted at least one year. Similarly, genetic distances increased as the number of extinction episodes or the number of failed reproductive episodes increased. These findings suggest that genetic distance is a more sensitive indicator of genetic change due to fragmentation, population bottlenecks and inbreeding.
Overall, our findings support the notion that lima beans populations are not in equilibrium. This results from complex demographic dynamics typical of this species, as they colonize ephemeral habitat and are likely to experience local extinction as environmental conditions change. In habitats where human activities such as agriculture, establishment of pasture and other practices maintain favorable environmental conditions for long periods, lima bean populations develop a genetic structure with a high level of population differentiation, and a within population substructure resulting from high levels of inbreeding and selfing and the occurrence of severe bottlenecks and founder effect.
Implications for conservation: The results of this study illustrate the importance of genetic monitoring for conservation biology. As pointed our by other authors (Schwartz et al. 2006), genetic monitoring can provide valuable information to help the decision making process leading to the establishment of adequate management strategies for the conservation of genetic diversity. Of particular relevance to this study are inbreeding coefficients, expected heterozygosity, temporal variances in allele frequencies and allele richness. Our data also support the notion that genetic and demographic data can be useful for identifying management units (see Palsbøll et al. 2006). Moreover, this study expands the uses of genetic data in management, as it examined levels of genetic differentiation over time, revealing that episodes of extinction and colonization may play an important role in determining spatial differentiation between population (Pannell and Dorkens, 2006).
Two of our findings have important implications for the adequate conservation of genetic resources of wild lima beans. First, small populations that undergo local extinction are often more likely to undergo changes in their genetic diversity than larger populations. These populations deserve special attention if they posses unique alleles, or other significant morphological or physiological traits, because they are likely to disappear in a random manner. However, because of within population structure, and other life history traits, large populations are not exempt from genetic change. Second, the integrity of the soil seed bank, as indicated by the production of seeds every year, also appears to have a significant effect on the conservation of genetic resources. Dormant seeds in the soil not only allow rapid recovery after disturbances, but also appear to maintain a good sample of the genetic diversity of the extant population.
Other authors have pointed out the role of severe genetic bottleneck in tropical plants (Aldrich and Hamrick 1998, Cespedes et al. 2003). They pointed out that such bottlenecks should be anticipated in the development of conservation strategies. However, long distance gene flow, via pollen or seed, may counter their negative effects (Mildén et al. 2006, Pannell and Dorken 2006). The genetic structure of wild lima beans in the Central valley of Costa Rica strongly suggests that flow (via pollen and seed) between populations is limited. However, more detailed studies are needed to elucidate its importance. We do not know the mechanisms for long distance dispersal in lima beans, but colonization events at new sites are not rare. We have recorded new populations of lima beans at sites where they have not been recorded since 1993 when we conducted our first large scale survey (Rocha et al. 1997, Barrantes 2003). Such dispersal events could explain, at least in part, the high levels of genetic differentiation between populations, and might represent an important factor in the dynamics of this species.
Acknowledgments
The authors thank M.E. Zaldivar, L. Castro, E. Castro, G. Aguilar, M. Artavia, and Ana Araya for advice, comments, and/or criticisms on a previous version of this manuscript. This work was supported by the Belgiums Direction Générale de la Coopération au Development and a University of Costa Rica grant (VI-111-91-223) to OJR.
Resumen
Las poblaciones de plantas pueden experimentar extinción local, y al mismo tiempo, pueden surgir a sus alrededores nuevas poblaciones. Algunas especies pueden colonizar el mismo sitio en múltiples ocasiones. Aquí examinamos el impacto de la extinción local y recolonización en la estructura genética de poblaciones silvestres del frijol lima (Phaseolus lunatus) en el valle Central de Costa Rica. Comparamos la diversidad genética de muestras tomadas en poblaciones, antes y después de la extinción, en 13 sitios, usando marcadores de microsatélite. Según los episodios de extinción durante los cinco años previos, clasificamos los sitios así: 1) poblaciones que han experimentado extinción por más de un año, y después han recolonizado (recolonizado), 2) poblaciones que no han experimentado extinción local (control), y 3) poblaciones que no han experimentado extinción local durante el estudio, pero fueron cortadas experimentalmente, simulando una extinción (experimental). Nuestros datos no mostraron una clara tendencia en la variación de las frecuencias alélicas, heterozigosidad, o número efectivo de alelos en y entre grupos de poblaciones. Los niveles de diferenciación genética entre muestras recolectadas en diferentes momentos en el mismo sitio fueron diferentes en los tres grupos de poblaciones. Los sitios recolonizados mostraron el mayor nivel de diferenciación genética (Fst = 0.2769), seguidos de los sitios control (Fst= 0.0576) y sitios experimentales (Fst= 0.0189). Obtuvimos resultados similares en la distancia genética Neis entre muestras (di,j = 0.1786, 0.0400, y 0.0037, respectivamente). Los cambios genéticos en los frijoles lima dependen de la duración y frecuencia de los episodios de extinción local. Las poblaciones "control" no están en equilibrio. Las implicaciones de estos resultados para el establecimiento de estrategias de conservación de los recursos genéticos de habas se encuentran en discusión.
Palabras clave: diversidad genética, metapoblación, diferenciación genética, banco de semillas del suelo, conservación in situ, frijol lima, Phaseolus lunatus, extinción local, recolonización, recursos genéticos.
Received 30-IV-2007. Corrected 30-X-2007. Accepted 21-IV-2008.
References
Aldrich P.R. & J.L. Hamrick. 1998. Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science. 281: 103-105. [ Links ]
Alvarez-Buylla, E.R., A.Chaos, D.Pinero & A. Garay . 1996. Demographic genetics of a pioneer tropical tree species: Patch dynamics, seed dispersal, and seed banks. Evolution. 50: 1155-1166. [ Links ]
Barrantes D. 2003. Efecto de las Extinciones Locales sobre la Estructura Genética de Poblaciones Silvestres de Frijol Lima (Phaseolus lunatus) en el Valle Central de Costa Rica. Masters thesis, SEP, University of Costa Rica, San José, Costa Rica. [ Links ]
Barton N.H. & M.G. Whitlock. 1997. The evolution of metapopulations. In I.A. Hanski , M.E. Gilpin (eds.). Metapopulation biology: ecology, genetics and evolution, Academic, San Diego, California, 183-214. [ Links ]
Baudoin J.P., O.J. Rocha, J. Degreef, A. Maquet & L. Guarino. 2004. Ecogeography, demography diversity and conservation of Phaseolus lunatus L. in the Central Valley of Costa Rica. Systematic and Ecogeographic studies on crop gene pools 12. International Plant Genetic Resources Institute, Rome, Italy. [ Links ]
Cespedes M., M.V. Gutierrez, N.M. Holbrooke & O.J. Rocha. 2003. Restoration of genetic diversity in the dry forest tree Swietenia macrophylla (Meliaceae) after pasture abandonment in Costa Rica. Mol. Ecol. 12: 3210-3212. [ Links ]
Cruzan M.B. 2001. Population size and fragmentation thresholds for the maintenance of genetic diversity in the herbaceous endemic Scutellaria Montana (Lamiaceae). Evolution 55: 1569-1580. [ Links ]
Degreef J., J.P. Baudoin & O.J. Rocha. 1997. Case studies on breeding systems and its consequences for germplasm conservation. 2. Demography of wild Lima bean populations in the Central Valley of Costa Rica. Genet. Resour. Crop. Ev. 44: 429-438. [ Links ]
Degreef J. 1998. Développement dun Model Démographique et Application à la Conservation In Situ des Populations Sauvages de Haricot de Lima (Phaseolus lunatus L.) dans la vallée Centrale du Costa Rica. Doctoral thesis, University of Agronomic Sciences, Gembloux, Belgium, 191 pp. [ Links ]
Degreef J., O.J. Rocha, T. Vanderborght & J.P. Baudoin. 2002. Soil seed bank and seed dormancy in wild populations of Lima bean (Fabaceae): considerations for in situ and ex situ conservation. Am. J. Bot. 89:1644-1650. [ Links ]
Ellstrand N.C. & D.R. Elam. 1993. Population genetic consequences of small population size: implications for plant conservation. Annu. Rev. Ecol. Syst. 24: 217-242. [ Links ]
Frankel O.H & M.E. Soulé. 1981. Conservation and evolution. Cambridge University, Cambridge. U.K. [ Links ]
Giles B.E & J, Goudet. 1997. Genetic differentiation in Silene dioica metapopulations: estimation of spatiotemporal effects in a successional plant species. Am. Nat. 149: 507-526 [ Links ]
Gillman M. 1997. Plant population ecology. In N. Maxted, B.V Ford-Lloyd, J.G Hawkes (eds.). Plant genetic conservation: the in situ approach. Chapman and Hall, London, England. 114-131. [ Links ]
Gliddon C. & J. Goudet. 1994. The genetic structure of metapopulations and conservation biology. In V. Loeschke, J. Tomiuk, SK Jain (eds.). Conserv. Genet. 107-114. [ Links ]
González D.O., Palacios N., Gallego C. & J. Tohme. 1995. Protocolos para marcadores moleculares. Unidad de Investigación en Biotecnología. Centro de Agricultura Tropical (CIAT). Colombia. [ Links ]
Hamrick J.L. & MJW Godt. 1989. Allozymes diversity in plant species. In AHD Brown, MT Clegg, AL Kahler, BS Weir (eds.). Plant Population Genetics, Breeding and Genetic Resources. Sinauer Associates, Sunderland, Massachusetts, USA. 43–63. [ Links ]
Hanski I. 1991. Single-species metapopulation dynamics: concepts, models and observations. Biol. J. Linn. Soc. 42:17–38. [ Links ]
Hanski I. 1999. Metapopulation ecology. Oxford Series in Ecology and Evolution. Oxford University, New York, USA. [ Links ]
Hanski I. & M. Gilpin. 1991. Metapopulation dynamics: brief history and conceptual domain. Biol. J. Linn. Soc. 42: 3-16. [ Links ]
Hanski I. & M.E. Gilpin. 1997. Metapopulation Biology: Ecology, Genetics and Evaluation. Academic, New York, USA. [ Links ]
Hanson L. 1991. Dispersal and connectivity in metapopulations. Biol. J. Linn. Soc. 42: 89-103. [ Links ]
Hardy O., S. Dubois, I. Zoro Bi & J.P. Baudoin. 1997. Gene dispersal and its consequences on the genetic structure of wild populations of Lima bean (Phaseolus lunatus) in Costa Rica. Plant Genetic Res. Newsl.109:1-6. [ Links ]
Harrison S. & J.F. Quinn. 1989. Correlated environments and the persistence of metapopulations. Oikos 56: 293-298. [ Links ]
Hartl D.L. & A.G. Clark. 1989. Principles of population genetic. Second Edition. Sinauer, Sunderland, Massachusetts, USA. [ Links ]
Husband B.C. & S.C.H. Barrett. 1996. A metapopulation perspective in plant population biology. J. Ecol. 84: 461-469. [ Links ]
Kalisz S., Horth L. & M.A. McPeek. 1997. Fragmentation and the Role of Seed Banks in Promoting Persistence of Collinsia verna in Isolated Populations. In MW Schwartz (ed.). Conservation in Highly Fragmented Landscapes. Chapman and Hall, New York, USA. 268-312. [ Links ]
Levins R. 1969. Some demographic and genetic consequences of environmental heterogeneity for biological control. B. Entomol. 15: 237-240. [ Links ]
Lodhi M.A., Ye G.N., Weeden N.F. & B.I. Reisch. 1994. A simple and efficient method for DNA extraction from grapevine cultivars and vitis species. Plant. Mol. Biol. Rep. 12: 6-13. [ Links ]
Maquet A., I. Zoro Bi, O.J. Rocha & J.P. Baudoin. 1996. Case studies on breeding systems and its consequences for germplasm conservation. 1. Isozyme diversity in wild Lima bean populations in the Central valley of Costa Rica. Genet. Resour. Crop. Ev. 43: 309-318. [ Links ]
Maquet A., I. Zoro Bi, M. Delvaux, B. Wathelet & J.P. Baudoin. 1997. Genetic structure of a Lima bean base collection using allozyme markers. Theor. Appl. Genetics 95: 980-991. [ Links ]
McCauley DE. 1993. Evolution in metapopulations with frequent local extinction and recolonization. In D Futuyma & J. Antonovics (eds.). Oxford Surveys in Evolutionary Biology, Volume 9 Oxford University, Oxford, UK. 109-134. [ Links ]
Menges E.S. 1990. Population viability analysis of an endangered plant. Conserv. Biol. 4: 52-62. [ Links ]
Menges E.S., C.V. Hawkes, W.J. Platt & K.R. Peet. 1998. Interactive effects of fire and microhabitat on plants of Florida scrub. Ecol. Appl. 8: 935-946. [ Links ]
Mildén M., Z. Münzbergová, T. Heven & Ehrlén. 2006. Metapopulation dynamics of a perennial plant, Succisa pratensis, in an agricultural landscape. Ecol. Model. 199: 464-475. [ Links ]
Nason J.D., P.R. Aldrich & J.L. Hamrick. 1997. Dispersal and the dynamics of genetic structure in fragmented tropical tree populations. In Tropical forest remnants: ecology management and conservation of fragmented communities. Laurance WF, Bierrgaad RO Jr (eds.). University of Chicago, Chicago, Illinois, USA. 304-320. [ Links ]
Nei M. 1973. Analysis of genetic diversity in subdivided populations. P. Natl. Acad. Sci. USA. 70: 33213323. [ Links ]
Newman D. & D. Pilson. 1997. Increased Probability of Extinction Due to Decreased Genetic Effective Population Size: Experimental Populations of Clarkia pulchella. Evolution. 51: 354-362. [ Links ]
Olivieri I., D. Couvert & P.H. Gouyon. 1990. The genetics of transient populations: research at the metapopulation level. Trends Ecol. Evol. 5: 207–210. [ Links ]
Ouborg N.J., Y. Piquot & J.M. Van Groenendael. 1999 Population genetics, molecular markers and the study of dispersal in plants. J. Ecol. 87: 551-568. [ Links ]
Palsbøll P.J., M. Bérubé & A.W. Allendorf. 2006. Identification of management units using population genetic data. Trends Ecol. Evol. 22: 11-16. [ Links ]
Pannell J.R. & B. Charlesworth. 2000. Effects of metapopulation processes on the measures of genetic diversity. Phil. Trans. Roy. Soc. London Series B 355: 1851-1854. [ Links ]
Pannell J.R. & M.E. Dorken. 2006. Colonization as a commoen denominator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landscape Ecol. 21: 837-848. [ Links ]
Poschlod P. 1996. The metapopulation concept-a consideration from the plant ecological viewpoint. Ziets. Okol. Naturschultz. 5: 161-185. [ Links ]
Rocha O.J., G. Macaya & J.P. Baudoin. 1997. Causes of local extintion and recolonization, determined by 3 years of monitoring wild populations of Phaseolus lunatus L. in the Central Valley of Costa Rica. Plant Genetic Res. Newsl. 112: 44-48. [ Links ]
Rocha O.J., Degreef J., D. Barrantes, E. Castro, G. Macaya & L. Guarino. 2002. Metapopulation dynamics of Lima bean (Phaseolus lunatus L.) in the Central Valley of Costa Rica. In JMMM Engels, V Ramanatha, AHD Brown, MT Jackson (eds.). Managing Plant Genetic Diversity, CABI Publishing, Wallingford, UK, 205–215. [ Links ]
Rocha O.J. & G. Aguilar. 2001. Reproductive biology of the dry forest tree Enterolobium cyclocarpum (Guanacaste) in Costa Rica: a comparison between trees left in pastures and trees in continuous forest. Am. J. Bot. 88: 1607-1614. [ Links ]
Schwartz M.K., Luikart Gordon & R.S. Waples. 2006. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22: 25-33. [ Links ]
Silvertown J. 1991. Dorothys dilemma and the unification of plant population biology. Trends Ecol. Evol. 6: 346-348. [ Links ]
Slatkin M. 1977. Gene flow and genetic drift in a species subject to frequent local extinctions. Theor. Popul. Biol. 12: 253-262. [ Links ]
Slatkin M. 1987. Gene flow and the geographic structure of natural population. Science . 236:87–92. [ Links ]
Sork V.I., J. Nason, D.R. Campbell & J.F. Fernández. 1999. Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol. Evol. 14: 219-224. [ Links ]
Valverde T. & J. Silvertown. 1997. A metapopulation model for Primula vulgaris, a temperate forest under-storey herb. J. Ecol. 85: 193–210. [ Links ]
Wade M.J. & D.E. McCauley. 1988. Extinction and recolonization: their effects on the genetic differentiation of local populations. Evolution. 42: 995–1005. [ Links ]
Whitlock MC & D.E. McCauley. 1990. Some population genetic consequences of colony formation and extinction genetic correlations within founding groups. Evolution. 44: 1717-1724. [ Links ]
White G. & H. Boshier. 2000. Fragmentation in Central American dry forests: impacts on Swietenia humilis (Meliaceae). In: A.G. Young & G.M. Clarke (eds.). Genetics demography and viability of fragmented populations, Cambridge University, UK, 293-311. [ Links ]
Wier B.S. & C.C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution. 38: 1358-1370. [ Links ]
Yeh F.C., R. Yang & T. Boyle. 1999. POPGENE version 1.31: Microsoft Windows-based Freeware for Population Genetic Analysis. University of Alberta, Edmonton, Canada. [ Links ]
Yu K., SJ Park, V Poysa & P. Gepts. 2000. Integration of Simple Sequence Repeat (SSR) Markers into a Molecular Linkage Map of Common Bean (Phaseolus vulgaris L.). J. Hered. 91: 429-434 [ Links ]
Zoro Bi I., A. Maquet & J.P. Baudoin. 1997. Spatial patterns of allozyme variants within three wild populations of Phaseolus lunatus L. from the Central Valley of Costa Rica. Belgium. Trends Ecol. Evol. 129: 149-155. [ Links ]
Zoro Bi I. 1999. Variabilité Génétique des Populations Sauvages de Phaseolus lunatus L. dans la vallée Centrale du Costa Rica et ses Implications dans la Mise au Point dune Stratégie de Conservation In Situ. Doctoral thesis, Faculté Universitaire des Sciences Agronomiques, Gembloux, Belgium, 194 pp. [ Links ]
Zoro Bi I., A. Maquet & J.P. Baudoin. 2003. Population genetic structure of wild Phaseolus lunatus (Fabaceae), with special reference to population sizes. American. J Bot. 90: 897-904 [ Links ]
Zoro Bi I., A Maquet & J.P. Baudoin. 2005. Mating system of wild Phaseolus lunatus L. and its relationship to population size. J. Hered. 94: 153-158. [ Links ]