Introduction
Taeniasis and cysticercosis are Neglected Tropical Diseases (NTD) produced by Taenia solium, T. saginata, and T. asiatica. The adult tapeworms are found in the human intestine causing taeniasis. The metacestodes or cysticerci (larval stages) cause cysticercosis; cysticerci of T. saginata produce bovine cysticercosis; cysticerci of T. asiatica develop porcine cysticercosis; and cysticerci of T. solium cause cysticercosis in pigs and humans. When cysticerci invade the central nervous system (CNS) produce neurocysticercosis (NCC) that is the most frequent parasitic infection of the human CNS. Taeniasis usually causes few symptoms, but NCC can be fatal, depending on the cysticerci location, number, and stage, and immune response of the host (Ferrer and Gárate, 2014; PAHO/ WHO, 2019; WHO, 2021).
T. solium and T. saginata show a wide geographical distribution, while T. asiatica has been described in Southeast Asia. Taeniasis remains to cause health problems and losses in the livestock industry from areas where these parasites are endemic; it also has affected non-endemic areas due to travels and migrations. NCC is common in many countries of Africa, Asia, and Latin America, specifically in communities with low socio-economic conditions and poor sanitation-hygienic practices. Useful tools for diagnosis, treatment, and protection are required to prevent, control, and possibly eliminate these diseases (Ferrer and Gárate, 2014; PAHO/WHO, 2019; WHO, 2021).
The characterization of genes is essential for the knowledge of parasite biology, the understanding of the parasite-host relationship, and the identification of possible targets to improve diagnostic techniques, treatment, and protection. A common non-translatable RNA sequence was discovered at the 5` end of mRNA encoding surface glycoproteins of Trypanosoma brucei, which was named Spliced Leader (SL) (Sather & Agabian, 1985). This molecule is inserted into the pre-mRNAs by trans-splicing, forming different mature mRNAs that contain a common 5 'end. This mechanism has been described in a great diversity of organisms, including nematodes, trematodes, and cestodes (Krchňáková et al., 2017). This mechanism of processing of mRNAs occurs in T. solium, like other parasites (Brehm et al., 2000, 2002; Garrido et al., 2012, 2015). The fraction of trans-spliced mRNAs varies between species. Although all the RNA of the genus Trypanosoma undergoes this post-transcriptional modification, in most of the transcripts of the other genera do not undergo trans-splicing, and the characteristics of the immature mRNA molecules that undergo this modification are unknown (Garrido et al., 2015; Krchňáková et al., 2017). The cloning strategy using the known SL sequence and sequences from a vector have allowed the cloning of complementary DNAs (cDNAs) from expression libraries of T. solium metacestode (Brehm et al., 2002; Garrido et al., 2012). In this work, we performed the molecular and bioinformatic characterization of three cDNAs (TsTF10, TsAAP8, and TsrGAP8), obtained by spliced leader-PCR screening of a Taenia solium cDNA library.
Methodology
The type of research was descriptive with a quantitative approach. The three cDNA were obtained by spliced leader-PCR screening of a Taenia solium cDNA library according to the protocol described by Brehm et al., (2002) and Garrido et al., (2012).
The primers TSSL-DW2 (5´-GGTCCCTTACCTTGCAATTTTGT-3´) and ZAP-3´UP (5´-GTAATACGACTCACTATAGGG-3´) were used to hybridize with the sequence SL and with a sequence of the vector, respectively (Brehm et al., 2002). The size of the cDNAs was determined by PCR. cDNAs products of different sizes were obtained, which were cloned into a pGEM-T-easy® plasmid and were sequenced following the protocol described by Garrido et al., (2015). The sequences of the three cDNAs were compared with the sequences in the nucleic acid and protein databases (GenBank, EMBL) and analyzed by bioinformatic programs.
Analysis of the nucleotide sequence and prediction of amino acid sequences were performed with the EditSeq program from DNAstar (Lasergene®, Madison, USA). Similarities were analyzed in the nucleic acid and protein databases (GenBank, EMBL) by BLAST (Boratyn et al., 2019). Other analyses of sequences were performed using CDD-Search of the National Center for Biotechnology Information (NCBI) (Lu et al., 2020), Interpro of the European Bioinformatics Institute (EBI) (Mitchell et al., 2019), Motif scan, and ExPASy (Expert Protein Analysis System) of the Proteomics Server from Swiss Institute of Bioinformatics (SIB) (Artimo et al., 2012). Epitopes B prediction was studied using the Protean program from DNAstar (Lasergene®, Madison, USA), and the BcePred server (Prediction of continuous B-cell epitope in antigenic sequences using physico-chemical properties) (Saha et al., 2005).
Analysis and Results
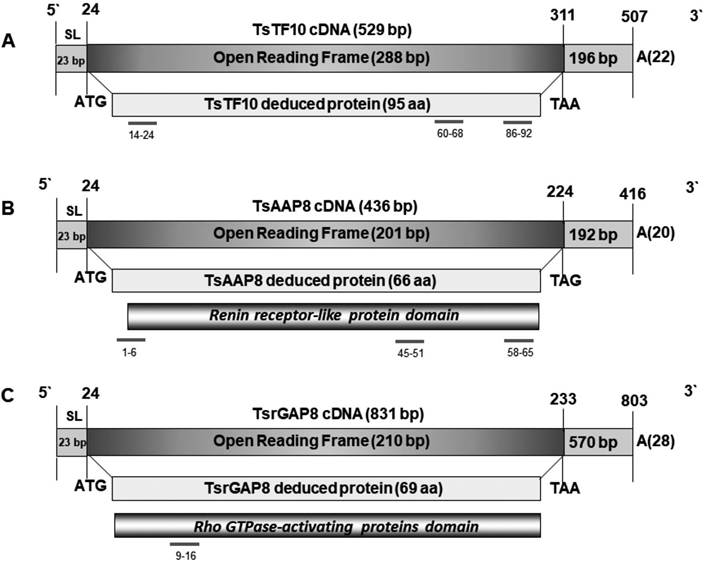
The results showed that the complete sequence of TsTF10 cDNA was a fragment of 529-bp with an open reading frame (ORF) of 288-bp that coded for a peptide of 95 amino acids, with a molecular mass of approximately 9.7 kDa, and an isoelectric point of 3.7. The ORF was preceded by a 5` spliced leader of 23-bp, and followed by a 3` untranslated region of 196-bp, and a 22-bp poly (A)+ tail. The deduced amino acid sequence showed four possible phosphorylation sites, and a Phenylalanine and histidine ammonia-lyase motif (PAL repeat, pFam PF00221). Regarding its possible immunogenicity, three B epitopes could be predicted in the molecule (14VVASSAGSSDE24, 60VQTTASSEE68, and 86QKLEEPS92) (Fig. 1A). The sequence data were submitted to the GenBank and are available with the accession number MW448478. This sequence showed high identity with the unnamed protein product of Taenia asiatica (VDK34809.1), with nuclear transcription factor γ gamma of other species, mainly from Echinococcus granulosus (XP_024354733.1) and Echinococcus multilocularis (CDS37349.1), ADP-ribosylation factor-like protein 2 of E. granulosus (XP_024351600.1), and unnamed protein product of Hymenolepis diminuta (VUZ44994.1) (Table 1).
The full sequence of TsAAP8 cDNA was a fragment of 436-bp, with an ORF of 201-bp that coded for a protein of 66 amino acids, with a molecular mass of approximately 7.5 kDa, and an isoelectric point of 4.3. The ORF was preceded by a 5` spliced leader of 23-bp, and followed by a 3` untranslated region of 192-bp, and a 20-bp poly (A)+ tail. The inferred amino acid sequence exhibited a probable N-glycosylation site, a casein kinase II phosphorylation site, a transmembrane section in the central part of the molecule (20-47 amino acids), and a characteristic domain of Renin receptor-like protein (Renin_r) (pFam PF07850, Interprot IPR012493).
Considering its possible immunogenicity, three B epitopes could be predicted in the molecule (1MANSSL6, 45WNMDPGR51, and 58LSVTKPKS65) (Fig. 1B). The sequence data were registered in GenBank under accession number MW452936. This sequence showed high percent identity with putative vacuolar ATPase membrane sector associated protein of Taenia solium (CAD21533.1), the unnamed protein product of T. asiatica (VDK22591.1), Intersectin-1 of Echinococcus granulosus (XP_024348633.1), dynamin associated protein of E. multilocularis (CDI96500.1), and unnamed protein product of H. diminuta (VDL61899.1) (Table 1).
TsrGAP8 cDNA showed an 831-bp nucleotide sequence, with an ORF of 210bp that coded for a peptide of 69 amino acids, with a molecular mass of approximately 7.7 kDa, and an isoelectric point of 7.2. The ORF was preceded by a 5` spliced leader of 23-bp, and followed by a 3` untranslated region of 570-bp, and a 28-bp poly (A)+ tail. The assumed amino acid sequence showed six potential phosphorylation sites, and a Rho GTPase-activating proteins domain (pFam PF00620, Interprot IPR000198). Regarding its possible immunogenicity, only a B epitope (9DHLKRITS16) could be predicted in the sequence (Fig. 1C). The sequence data were submitted to the GenBank and is available with the accession number MT707920. This sequence was highly similar to; Rho GTPase activating protein of Echinococcus granulosus (CDS19130.1), Rho GTPase activating protein of E. multilocularis (CDS37180.1), the unnamed protein product of Taenia asiatica (VDK32116.1), the unnamed protein product of Hymenolepis diminuta (VUZ54861.1), and Rho GTPase activating protein of H. microstoma (CDS27177.2) (Table 1).
Table 1 Similarities between the TsTF10, TsAAP8, and TsrGAP8 cDNAs sequence and other GenBank sequences by BLAST.
| cDNA | Similar molecules | % identity (aa) |
|---|---|---|
| Unnamed protein product of Taenia asiatica (VDK34809.1) | 97.9 | |
| Nuclear transcription factor gamma of Echinococcus granulosus | 85.1 | |
| (CDS19297.1)* | ||
| TsTF10 | Nuclear transcription factor gamma of Echinococcus multilocularis | 83.0 |
| (CDS37349.1)* | ||
| ADP-ribosylation factor-like protein 2 of Echinococcus granulosus | 82.6 | |
| (XP_024351600.1)* | ||
| Unnamed protein product of Hymenolepis diminuta (VUZ44994.1)* | 57.3 | |
| Putative vacuolar ATPase membrane sector associated protein of Taenia solium (CAD21533.1)* | 98.5 | |
| Unnamed protein product of Taenia asiatica (VDK22591.1)* | 98.0 | |
| TsAAP8 | Intersectin-1 of Echinococcus granulosus (XP_024348633.1)* | 93.1 |
| Dynamin associated protein of Echinococcus multilocularis (CDI96500.1)* | 92.3 | |
| Unnamed protein product of Hymenolepis diminuta (VDL61899.1)* | 74.2 | |
| Rho GTPase activating protein of Echinococcus granulosus (CDS19130.1)* | 98.5 | |
| Rho GTPase activating protein of Echinococcus multilocularis | 98.5 | |
| (CDS37180.1)* | ||
| TsrGAP8 | Unnamed protein product of Taenia asiatica (VDK32116.1)* | 98.5 |
| Unnamed protein product of Hymenolepis diminuta (VUZ54861.1)* | 89.7 | |
| Rho GTPase activating protein of Hymenolepis microstoma (CDS27177.2)* | 89.7 |
*GenBank accession number, (aa) amino acids
Note: derived from research.
Note: derived from research.
Figure 1 Schematic representation of the three cDNAs analyzed of Taenia solium. (A) TsTF10, (B) TsAAP8, (C) TsrGAP8. The bars below deduced proteins represent the domains predicted in the sequences. The short grey lines with below numbers represent the positions of the B epitopes found. (bp)base pairs (aa)amino acids.
Discussion
In this work we have characterized three new molecules of T. solium metacestode, with respect to the potential function of these T. solium genes. Taking into account the high sequence identity to similar molecules of related helminths (Taenia asiatica, Echinococcus granulosus, E. multilocularis and Hymenolepis diminuta) and the functional domains found, the TsTF10, TsAAP8, and TsrGAP8 genes of Taenia might act as a nuclear transcription factor gamma, a putative vacuolar ATPase membrane sector associated protein, and a Rho GTPase activating protein, respectively.
Transcription factors (TFs) are DNA-binding proteins that regulate gene expression, and they have decisive roles in the control of cellular performance. Although there is significant progress, we even now have an incomplete understanding of how genomic and epigenomic information guides gene expression through specific transcription factors (Chen & Franklin, 2021). TsTF10 gene could mean T. solium specific transcription factors since no significant similarity was found with sequences homologous from human or other species (only with cestodes parasites), therefore, it could be a therapeutic or protection target.
The renin receptor-like protein domain (Renin_r) corresponds to a similar region of the human renin receptor that bears a putative transmembrane spanning segment and is involved in intracellular signal transduction. On the other hand, this proteins family (bear Renin_r domain) also includes ATPase H(+)-transporting accessory protein 2, which is known as ATP6AP2, and renin homolog receptor. ATP6AP2 protein serves as a receptor in the two-dimensional cell polarity (PCP), as well as being implicated in the assemblage of the proton-transporting vacuolar (V)-ATPase protein pump. The vacuolar-type H+-ATPase (V-ATPase) is a multi-subunit enzyme composed of a peripheral V1 complex, responsible for the hydrolysis of ATP, and an integral V0 complex responsible for the transport of protons crosswise endomembrane or plasma membranes. They are found in the endomembrane (endosomes, lysosomes, and secretory vesicles) of all eukaryotes and in the plasma membranes of many eukaryotic cells (Liu et al., 2006). Consequently, the TsAAP8 gene seems to code for an integral component of membrane associated to (V)-ATPase protein pump of the parasite and has a signaling receptor activity. Moreover, TsAAP8 could be a T. solium specific vacuolar ATPase membrane sector associated protein since the identity with the human and pig homologous is about 34-35%; therefore, this sequence difference (65%) (molecule fragment) seems to mean a therapeutic or protection targets.
The Rho GTPase-activating proteins domain is characteristic of a proteins family that act as molecular switches, hence, it remains active in its GTP-bound form but inactive when it is bound to GDP. The Rho family of small G proteins activates effectors involved in a wide variety of developmental processes, including the regulation of cytoskeleton formation, cell proliferation, and the JNK signaling pathway. G proteins usually have low essential GTPase hydrolytic activity. However, there are specific groups of GAPs that improve the proportion of GTP hydrolysis by some orders of magnitude. The RhoGAPs are one of the main classes of controllers of Rho G proteins. Rho GTPase-activating proteins catalyze the change of active GTP-bound Rho to inactive GDP-bound Rho by increasing GTP hydrolysis. In cells, Rho activity regulates the actin cytoskeleton organization and the actomyosin II contractility (Hanley et al., 2020). TsrGAP8 showed about 33-52% of identity with human and pig homologous isoforms and it could be a therapeutic target due to this sequence difference (48-67%).
In the GenBank, there is a putative vacuolar ATPase membrane sector associated protein of T. solium, with 98.5% of identity to TsAAP8, which have been described by Brehm et al. (2002), but it has not been characterized. TsAAP8 protein could be an isoform due to the slight difference between amino acid sequences (1.5%). Few B epitopes were predicted with high probability from three molecules of a T. solium cDNA library. Even though they do not seem to be as other antigenic molecules, their evaluation as possible candidates might be relevant for diagnosis and protection in cysticercosis. The principle that housekeeping and structural proteins do not work well as antigens has long been sustained. Most of housekeeping proteins are inner proteins and are shown to the immune system in the late stages of infection, after parasites lysis. Furthermore, the fact that most of these proteins are highly conserved, sustains the idea that such proteins should not be good antigens. However, some studies with this type of proteins showed that they are able to generate a robust immune response in the host (Cook et al., 2004; Morillo et al., 2020). TsTF10 and TsAAP8 proteins could be used in the diagnosis and/or protection studies since more B epitopes were predicted.
Although there is a Taenia solium genome project (Tsai et al., 2013) and the sequences of all the genes are there, there are no published studies on the molecules TsTF10 and TsrGAP8. At the National Autonomous University of Mexico, a consortium of numerous laboratories carried out a sequencing project for T. solium. They have reported that most of the expressed sequence tags (ESTs) of T. solium are related to gene regulation, and signal transduction. Other important functions are cytoskeleton, housekeeping, cell division, metabolism, hormone response, vacuolar transport, proteases, and extracellular matrix activities (Tsai et al., 2013). These functions and activities are essential to the adaptation to parasitism which is according to the possible functions of the three molecules described in this work.
The characterization and analysis of these sequences and the prediction of their possible usefulness as antigens, vaccines, or therapeutic targets, contribute to the designing and planning of future studies.
Conclusions
The spliced leader-PCR screening of a T. solium cDNA library is a helpful strategy to obtain molecules from cDNA libraries. The TsTF10, TsAAP8, and TsrGAP8 genes of Taenia could act as a nuclear transcription factor gamma, a putative vacuolar ATPase membrane sector associated protein, and a Rho GTPase activating protein, respectively. The characterization and analysis of these sequences and the prediction of their possible usefulness help to design and plan future studies.
Funding
This study received financial support from Project CDCH-UC-010610-2008, Universidad de Carabobo, Venezuela.















