A better understanding of the generation of antimicrobial resistance (AMR) and its presence in poorly investigated natural environments can lead to combating this global health challenge. AMR is constantly increasing due to the mismanagement of antimicrobial compounds (Lee & Lin, 2003) and continues to be reported worldwide, expanding into natural environments as a result of urbanization and vector spread. In fact, chloramphenicol-resistant strains have even been detected in surface waters of isolated cave microbiomes in Mexico (Bhullar et al., 2012).
Water catchments stored in the natural cavities of plants (phytotelms), such as bromeliad tanks, are commonly used as bioindicators. Their physicochemical and geochemical characteristics, as well as the communities of macro- and micro-organisms, and their interactions, make them suitable matrices for evaluating ecological interactions, carbon and nitrogen cycles, and environmental stresses (Benavides-Gordillo et al., 2019). Moreover, the bacterial communities within bromeliad tanks are unique, and their bacterial abundance remains stable even during drought or increased rainfall events (Rodríguez-Pérez et al., 2018; Benavides-Gordillo et al., 2019).
To date, no studies have investigated AMR in phytotelm samples from a tropical cloud forest in Costa Rica. The closest relevant research was conducted by Brighigna et al. (1997), who successfully used Tillandsia caput-medusae (Bromeliaceae) as a biomonitor of air pollution by lead, copper, and cadmium. Therefore, the aim of this research was to assess the potential of phytotelms as a matrix for detecting antibiotic-resistant bacterial strains in the deep understory of a tropical cloud forest and a nearby rural town in Costa Rica. We employed a simple, inexpensive, and rapid method to evaluate these bacterial communities and determine the extent to which antibiotics have penetrated the forest before proceeding with molecular identification. Additionally, we conducted a text mining analysis to investigate the current information available on "antibiotic resistance in forests" and determine the direction of existing studies. We isolated six gram-negative bacterial strains from the forest and four from the urban site. Only one strain was susceptible to all antibiotics and three strains were multi-resistant to different antibiotics (>3) in each site (Fig. 1A, B). Congruently, NMDS and PERMANOVA revealed that both locations had a similar response to antibiotic resistance (F(1,13) = 1,02, P>0,05, Fig. 1C).
We identified 71 articles that aligned with our search conducted on PubMed.gov. Among this limited number of articles, there has been a significant and rapid increase in interest in studying the presence of antibiotic resistance in forests over the past five years (Figure 2A). The oldest was an article on the ecology of aquatic bacteria and multiple antibiotic resistance (Ogan & Nwiika, 1993), but it was not until the years 2018-2021 that the interest in this topic experienced a notable upturn. Figure 2B illustrates the recurring concepts associated with forests, including genes, soil microbiology, environmental monitoring, agriculture, land use, rivers, antibiotic resistance genes, and microbiota. We raise the hypothesis that the AMR could be introduced into the forest by the exposition of antimicrobial resistance genes (ARGs), resistant bacteria, or antibiotic molecules transported by different dissemination pathways such as wind, precipitation, air pollution, or as via fine particulate matter (PM 2.5) contained in the atmosphere (Xie et al., 2018; Zhu et al., 2021). Segawa et al. (2013), revealed the presence of resistance genes due to clinical tests in polar snow and glaciers, where humans have not intervened, and argue that these were spread by bacteria through the air. The phytotelm is a complex and small ecological habitat with several community interactions, where bacteria produce their antimicrobial compounds to compete for space and nutrients, leafding to the generation of natural resistance. In addition, bacteria can mutate, adapt, and transfer multi-resistance genes, which may enhance the mechanisms of AMR gene transfection in this small niche (von Wintersdorff et al., 2016).
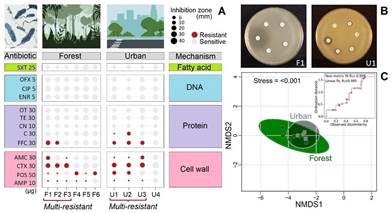
Fig. 1 A) Test of gram-negative bacterial antibiotic resistance categorized by type antibiotic and inhibition mechanism for forest and urban sites. The size of the circles shows the diameter of the bacterial growth inhibition halo. Bacteria with three or more resistant antibiotics are designated as multi-resistant. B) Illustration of two antibiograms. C) Non-metric multi-dimensional scaling (NMDS, presence/absence) of bacterial antibiotic resistance in phytotelm for forest and urban sites. Antibiotic: AMC- Amoxicillin/Clavulanic Acid, AMP-Ampicillin, C-Chloramphenicol, CIP-Ciprofloxacin, CN-Gentamicin, CTX-Cefotaxime, ENR-Enrofloxacin, FFC-Florfenicol, FOS-Fosfomycin, OFX-Ofloxacin, OT- Oxytetracycline, SXT-Trimethoprim/Sulfamethoxazole, and TE-Tetracycline
This is the first study to attempt to prove the presence of bacterial strains with some degree of antibiotic resistance in phytotelms from the deep understory of a tropical cloud forest from Costa Rica. The potential dangers of the emergence of antibiotic resistance in natural ecosystems are uncertain. Efforts to determine the impact of antibiotic resistance on the human population estimated that deaths from antimicrobial resistance could rise from about 700,000 a year to about 10 million deaths per year by 2050 (O’Neill, 2014), despite criticism from Kraker et al., (2016) and NOAH (2016). Although antibiotic resistance is a natural process, the use of antimicrobials has increased significantly in recent decades, by exposing bacteria to greater numbers and increased concentration, which increases their chances of developing resistance.
Our results open the possibility of using the phytotelm as a convenient and practical matrix for detecting AMR dissemination. The bacterial communities within these water catchments exhibit remarkable stability and resistance to fluctuations in water volume. This unique characteristic makes them an ideal matrix for assessing long-term environmental changes, distinguishing them from other commonly used samples. Additionally, this approach enables simultaneous monitoring of large protected forest areas, offering a fast and efficient tool.
Further research is necessary to determine whether AMR is primarily influenced by anthropogenic factors or occurs through natural mechanisms of bacterial AMR generation. While bacterial antibiotic susceptibility tests using the disk diffusion technique are a cost-effective, easy, and rapid method for identifying resistant and multi-resistant bacteria, they do not address antibiotic resistance genes (ARGs) or resistance in non-cultivable bacteria. However, the detection of traces of antibiotic compounds in the phytotelm can complement this method.
Lastly, it is crucial to consider how ecological and physicochemical factors can influence AMR patterns in different geographical locations where samples are collected.
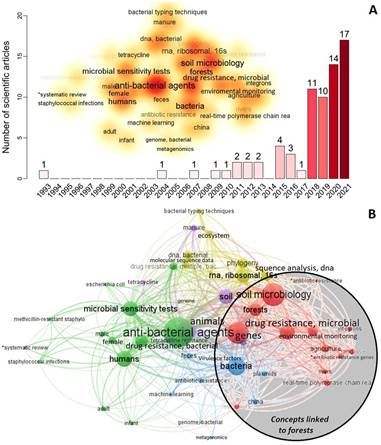
Fig. 2 A) The number of scientific articles about bacterial antibiotic resistance in forests, 1993-2020 (n = 71). Word cloud is the analysis of the frequency of the keywords of all the articles with co-occurrences greater than three words. B) Network analyses keywords of all the scientific articles with co-occurrences greater than three words












 uBio
uBio 


