Introduction
The use of DNA barcoding with COI sequences is proposed as a highly efficient identification tool for a diverse range of marine invertebrates. Given that interspecific divergence gaps tend to be wider than intraspecific divergences, divergence thresholds demonstrate the capability to effectively distinguish recently diverged groups, leading to more accurate putative species assignments in taxonomic research compared to morphological approaches, especially for cryptic complexes (Laakmann et al., 2016; Layton et al., 2016). Therefore, molecular taxonomy is a valuable alternative for solving conflicts related to the application of morphology-based taxonomic diagnosis, particularly when open-access barcode reference libraries are available. There are 23 297 echinoderm COI entrances in GenBank and 37 267 public records in BOLD for at least 2 115 echinoderm species (verified 18 Nov. 2023). However, only few studies have tested the accuracy of public sequence repositories in DNA identification (Laakmann et al., 2016; Meiklejohn et al., 2019) and none have explored it for Central American echinoderm molecular taxonomy.
Many Central American countries share a common origin in echinoderm research, marked initially by North American and European expeditions in the late nineteenth century, leading to the description of the majority of Central American echinoderm species. Subsequently, there was an increase in investigations conducted by regional scientists or research institutes in the second half of the twentieth century (Alvarado et al., 2013; Alvarado & Fabregat-Malé, 2021). Historically, samples have been kept in foreign museums, limiting access to valuable vouchers and type material for local scientists (Alvarado et al., 2017). Nevertheless, the Colección Nacional de Equinodermos Ma. Elena Caso Muñoz at Instituto de Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México, hosts one of the largest specimen collections in Latin America (Alvarado & Solís-Marín, 2013). Furthermore, the Museo de Zoología (MZ-UCR) at the Centro de Investigación en Biodiversidad y Ecología Tropical (CIBET, Universidad de Costa Rica) has consistently expanded its echinoderm collection, facilitating local scientists’ access to more in-depth voucher scrutinizing and creating opportunities for developing collaborative networks in regional integrative research (Cortés & Joyce, 2020; Kress, 2014; Miralles et al., 2020; Schilthuizen et al., 2015).
While there have been ongoing efforts in Central American echinoderm research with a consistent focus on ecological and taxonomic issues, the exploration of related areas as molecular taxonomy has been limited (Alvarado et al., 2013; Alvarado & Fabregat-Malé, 2021; Varela-Sánchez et al., 2020). Since GenBank and the Barcode of Life Data System (BOLD) are the main molecular public repositories, we tested their accuracy and reliability for COI echinoderm sequences identification at genus and species level (i.e. barcoding). This study started the establishment of a validated and comprehensive echinoderm reference library, comprising morphological, and molecular data, as well as additional metadata for samples collected at Área de Conservación Guanacaste, Costa Rica (Cortés & Joyce, 2020). However, the biogeographic and bathymetric distribution of many of the present species extends the applicability of this library to Central American Pacific shallow waters (< 200 m depth) (Alvarado et al., 2010; Solís-Marín et al., 2013). We matched morphological identifications with the molecular species assignments (correct identifications or identification errors) as a reference method for database improvement in further taxonomic and systematic studies in the region.
Materials and methods
Sample preparations: Sampling and tissue extraction for 475 vouchers were conducted during the BioMar-ACG echinoderm surveys in 25 sample sites (Fig. 1). Different body parts were dissected for DNA extraction depending on the taxonomic group, generally dermis, podia, or arm tips were dissected and stored in absolute ethanol (96 %) on lysis plates, then frozen until its processing. Identifications were done by direct observation, using optic stereoscopes and light microscopes. Taxonomic identifications were based on proposed diagnostic characters from external and internal anatomy or skeletal elements (i.e. ossicles) sourced in taxonomic specialized literature (Borrero-Pérez & Vanegas-González, 2019, Borrero-Pérez & Vanegas-González, 2020; Granja-Fernández et al., 2014, Granja-Fernández et al, 2020; Lessios, 2005; Martín-Cao-Romero et al, 2017; Massin et al., 2002; Solís-Marín et al., 2009, Solís-Marín et al., 2014, Solís-Marín et al., 2020; Woo et al., 2015). The nomenclature for taxonomic designations follows the World Register of Marine Species (WoRMS Editorial Board, 2024). When identifications could not be established at the species level, provisional identifications were given to the lowest possible taxon.
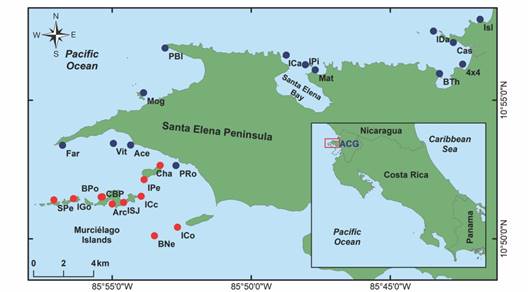
Fig. 1 Echinoderm sample sites in the BioMar-ACG project at Área de Conservación Guanacaste, north Pacific of Costa Rica. Peninsular sites (blue dots) and Archipelago sites (red dots) are shown. Modified from Chacón-Monge et al. (2021), published with permission.
Data collection: In order to assess the whole representation of the Central American shallow water Pacific echinoderm richness, we registered an updated list of species reported from the Central American Pacific coast (excluding Panama) from 0 to < 200 m depth (Appendix 1). The DNA extraction, amplification, and sequencing of COI fragments were conducted by the Center for Biodiversity Genomics, Guelph University, Canada (Cortés & Joyce, 2020). Throughout an automatized process developed and improved since the early 2000’s (Ivanova et al., 2006; deWaard et al., 2008; for detailed protocols). The obtained sequences were analyzed with the GenBank and BOLD identification tools.
DNA barcode analyses: The obtained sequences were individually searched with the Nucleotide Basic Local Alignment Search Tool (BLAST Best Match) in GenBank’s identification core (Altschul et al., 1990) and also compared with the Identification System (IDS) engine available in the Barcode of Life Data System, BOLD (Meiklejohn et al., 2019; Ratnasingham & Hebert, 2007). Setting the “standard nucleotide BLAST” and the “Public Record Barcode Database’’ for nucleic species identification respectively, each obtained sequence was requested under GenBank and BOLD built-in search tools. Query sequences were considered “Correctly identified” (C) if a record with the same taxonomic name had the best match statistic or highest percent identity, otherwise they were classified as an Identification error (IE) (Laakmann et al., 2016; Meiklejohn et al., 2019). Morphologically unsolved taxa were not included in identity comparisons, instead were labeled as No Apply (NA). Given our data nature, McNemar’s Chi-squared, Kruskal-Wallis’s and Mann-Whitney’s U tests were performed to prove the differences between these databases and their identification accuracy.
Results
Overall representativeness: About 466 echinoderm species have been registered in Central America, and around 364 (78 %) have been listed for Costa Rica (Alvarado & Fabregat-Malé, 2021; Alvarado et al., 2017; Alvarado et al., 2022; Chacón-Monge et al., 2021; Solís-Marín et al., 2013). We listed a total of 324 echinoderm species reported for Central American Pacific shallow waters (< 200 m) (Appendix 1), and about 236 (73 %) should be present in Costa Rica (Alvarado et al., 2017; Borrero-Pérez & Vanegas-González, 2020; Chacón-Monge et al., 2021; Granja-Fernández et al., 2020; Solís-Marín et al., 2020). Nevertheless, only 118 (36 %) and 110 (34%) of Central American echinoderm fauna were represented in the GenBank nucleotide search and BOLD public databases respectively, with sequences for only 96 shared species and sharing 192 absences (Appendix 1). Our database includes 348 COI samples successfully sequenced in the BioMar-ACG project for 44 morphology-based species in 325 sequences (Appendix 1), and 21 provisional identifications in seven taxa (Appendix 2). Representing only 19 % of Costa Rican and 14 % of Central American Pacific shallow water echinoderm species, but 37 % and 40 % of the total representativeness available for the region in each database (GenBank and BOLD).
Sequence recovery: In terms of sequence recovery, our study achieved a sequencing success rate of 73 %, resulting in 348 sequences out of 475 samples. Not too far from the 78 % obtained in a major Canadian echinoderm database (1 285 vouchers), where the sequencing failure was attributed to primer effectiveness (Layton et al., 2016). Sequence length ranges from 200 to 680 bp (average 629 bp). We proposed 325 solved and 21 unsolved morphology-based identities in 50 putative taxa (Appendix 2). All query sequences were identified using the GenBank nucleotide identification engine. There were 56 species proposed for 336 sequences and twelve provisional identities for three taxa, with correspondences from 81.2 % to 100 %. BOLD recovered 169 solved identifications in 22 taxa and one provisional identification, with correspondences from 97.06 % to 100 %. But 178 sequences were retrieved as unmatched terms (in 33 morphology-based taxa), last verification October 9th, 2023 (Appendix 2). The general taxonomic representativeness, success and numbers of proposed genera and species identities using each identification method is summarized by class (Appendix 3). Our study contributes to the inclusion of sequences from local vouchers in molecular databases. Specifically, we provide 84 sequences for eleven species not previously represented in GenBank and 65 sequences for nine species absent in BOLD (Appendix 2).
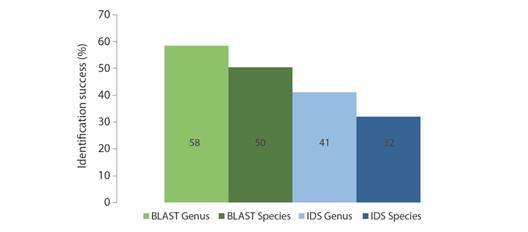
Barcoding identification: Although 39 of the putative morphology-based taxa were represented at GenBank COI database (in 265 seq.) and 41 morphology-based taxa were found at BOLD sequence database (in 283 seq.), not always the query sequence retrieved the expected identification. For genus and species level, identifications success differs (McNemar’s chi-squared = 32.84, df = 1, p < 0.01; McNemar’s chi-squared = 35.43, df = 1, p < 0.01) between identification engines, and was greater for GenBank at genus (Kruskal-Wallis = 52.44, df = 1, p < 0.01) and species level (Kruskal-Wallis = 41.625, df = 1, p < 0.01). According to the GenBank research engine, there were 175 sequences correctly identified and 152 identification errors, while in the BOLD public record barcode, there were 111 correct identifications and 216 identification errors (Table 1). Otherwise 21 Vouchers did not apply for this comparison, since they correspond to morphology-based unsolved identities (Appendix 2). Only 87 (24 %) and 128 (37 %) sequences were simultaneously correctly and incorrectly identified respectively (Appendix 2). Despite the identity percentage was higher for BOLD compared to GenBank (Mann-Whitney U = 2110, p < 0.01), the accuracy for genus and species identification using the BLAST Best Match was better than the obtained by the IDS engine, therefore GenBank outperforms BOLD identification at both taxonomic categories (Fig. 2).
Table 1 Classification of query sequences for Central American Pacific shallow water echinoderms using GenBank (BLAST) and BOLD (IDS) identification tools.
| ID | BLAST | IDS |
| Correct (C) | 175 | 111 |
| Identification Errors (IE) | 152 | 216 |
| NA | 21 | 21 |
| No match | 0 | 178 |
Discussion
Identification success: Low species identification success was not expected, given the relatively well representation of the proposed taxa related to Central American Pacific shallow water echinoderms in GenBank and BOLD libraries. It is also possible that the discriminatory power for species-level identification may be diminished on short sequences (< 430 bp), but similar results have been obtained using COI fragments of 650 bp and 130 bp for insect identification (Grzywacz et al., 2017).
Misidentifications: Identification discordances in both databases may obey specific parameters used in each search algorithm engine, and sequences available for barcoding under comparison (Layton et al., 2016). GenBank compares the query sequence to the nucleotide database, the Mega BLAST search tool (established by default) is optimized with similar sequences to search against all others using the command-line interface. While in BOLD, when public database records are chosen, a selection from the published projects section in BOLD (including all public COI records from BOLD and GenBank with a minimum sequence length of 500 bp) is automatically selected to compare the query sequence (Meiklejohn et al., 2019). Therefore, each requested sequence is compared against the appropriate group of barcode sequences according to the specific parameters for each database. General discordances can appear because of the specific evolutionary history of species, imperfect species delineation and overlooked diversity (“bad taxonomy” sensuEbach et al., 2006; Hörandl, 2007). In consequence, putative species are more reliable when supported by various delimitation methods and COI based techniques need to be evaluated under integrative taxonomy (Laakmann et al., 2016; Sonet et al., 2022).
Considering all identification errors and unmatched terms, after a consequent taxonomic revision and morphologic comprobation, we found 23 morphology based vouchers in four putative species erroneously identified or unsolved before the BLAST query (five Holothuria arenicola previously identified as H. impatiens; six H. impatiens identified as H. pardalis, two Ophioderma panamense identified as O. cf. panamense and ten O. hendleri identified as Ophioderma sp.). Seven of them were also correctly identified by BOLD in two species (five H. arenicola and two O. panamense, the other 16 were retrieved as unmatched terms. The rest of morphology-based unsolved taxa correspond to juvenile stages. Based on the molecular databases compared and employed analysis, the suggested misidentifications may correspond to real barcoding identification errors. Rather than possible morphology based errors, contrary to the expected rate of identification success using COI molecular DNA for echinoderm identification. We strongly recommend taking precautions when using public sequence databases to identify morphologically undetermined species of Central American Pacific shallow water echinoderms. With special awareness if they include not verified taxa, juvenile stages or information from “unpublished projects”, which might include unsolved taxonomy. In contexts where no prior taxonomic information about echinoderm samples is known, accurate and reliable interpretations of discordant identities may impose a challenge (Sonet et al., 2022).
Regional representativeness: This effort also marks a crucial step towards systematically evaluating the representativeness and accuracy of the GenBank and BOLD identification engines for echinoderm species in Central American Pacific shallow waters. Not only refining molecular identification tools, but also increasing the overall regional representativeness of COI sequences databases. This work is a first step in the DNA barcode library construction for Central American Pacific shallow water echinoderms, but additional echinoderm sample references are needed to improve the utility of DNA barcoding as an identification tool in the region.
Applicability: DNA barcoding is a complementary approach to biodiversity studies. It is possible to obtain robust results using these databases in combination, as well as to improve data sets for systematic purposes and further taxonomic research, by applying methods for putative species identification based on COI sequences for previously known taxonomic identities. Nevertheless, we do not recommend the use of molecular repositories as the main source for Central American Pacific shallow water echinoderm species identification. Echinoderm systematics could benefit from additional integrative taxonomy approaches and molecular data analysis only if it is keeping connection between voucher morphology and sequences. It directly highlights the importance of increasing the sequence’s representativeness for many common and rare species in global public scientific molecular repositories and holding vouchers in local natural history collections with corroborated taxonomy.
Ethical statement: the authors declare that they all agree with this publication and made significant contributions; that there is no conflict of interest of any kind; and that we followed all pertinent ethical and legal procedures and requirements. All financial sources are fully and clearly stated in the acknowledgments section. A signed document has been filed in the journal archives.
See supplementary material










 uBio
uBio