Introduction
The black soldier fly (BSF), Hermetia illucens (Linnaeus, 1758), is found worldwide in tropical and temperate regions (Kaya et al., 2021), mainly in manure and food waste dump grounds (Kim et al., 2008). In its larval state, it feeds on large amounts of decaying organic matter, which makes it very useful for accelerating composting processes, and able to convert low quality biomass into nutritional valuable proteins (Czekała et al., 2020; Diener et al., 2009). Furthermore, in its adult state, it is not considered a pest or a disease vector, making BFS larvae production safe and environmentally friendly (Joosten et al., 2020). These characteristics have increased BSF rearing and research on its microbiota, which is fundamental for the bioconversion process (Jiang et al., 2019).
The bacterial component of the BSF microbiome has been extensively studied, and a core bacterial community has been described (Cifuentes et al., 2022; IJdema et al., 2022; Jiang et al., 2019). Information about how diet source, insect developmental stage, and location in the digestive tract influence bacterial communities is also available (Bruno et al., 2019; Querejeta et al., 2023; Wynants et al., 2019). Fungal communities, on the other hand, have been much less studied and there is not enough data yet to elucidate if there is a core fungal community. In the last few years, there have been some studies about fungal composition using larvae reared under laboratory conditions in different substrates (Boccazzi et al., 2017; Klüber et al., 2022a; Klüber et al., 2022b; Tanga et al., 2021; Tegtmeier et al., 2021). However, information about the fungal composition of naturally occurring larvae is scarce, and most of the studies have been developed in subtropical or temperate regions, despite the high abundance of BSF in tropical regions (da Silva & Hesselberg, 2020).
Tropical regions, because of their intrinsic abiotic characteristics such as heat and high humidity, are recognized for concentrating the greatest species diversity on the planet (Brown, 2014), and fungal diversity is not an exception to this (Aime & Brearley, 2012; Boekhout et al., 2021). Analyzing mycobiomes on BSF could allow the identification of fungal species or strains with unique and important characteristics, such as the capacity to degrade complex substrates that can be useful in industry and bioremediation (Østergaard & Olsen, 2011). It might also contribute to the global problem of the identification of new antimicrobials (Gil-Rodríguez & Garcia-Gutierrez, 2021).
Urban compost, on the other side, has an increasing importance due to the rise in organic waste production in the last few decades. Fungi play a vital role in organic matter degradation and are responsible for compost maturation (Dehghani et al., 2012), thus characterizing mycobiomes related to composting process might help to make the process more efficient and faster (El Hayany et al., 2022). It can also help to discover thermotolerant microorganisms, as temperatures during composting can reach up to 70 °C due to the heat produced by exergonic aerobic reactions derived from microbial metabolism (Hemidat et al., 2018). Thermotolerant fungi have a special interest in industry because of the thermostability of their enzymes, such as thermostable carbohydrate active enzymes (CAZymes) (Di Piazza et al., 2020; Gabriel et al., 2021), as they could be used to improve industrial processes and have biotechnological applications (Cruz-Ramírez et al., 2012).
In this study, we characterize the fungal communities present on BFS larva gut naturally occurring during urban composting processes in a tropical region using Illumina high-throughput sequencing of the ITS region. Our aim is to identify the principal fungal groups and to compare them with previous reports under different conditions. This information can contribute to a better understanding of the ecology of this species, the composting microbial dynamics, and it can set a precedent for further research on the possible industrial and biotechnological applications of the microorganisms found.
Materials and methods
Sample collection and dissection: A total of five H. illucens larvae (two to three weeks old) (Fig. 1) were collected from household compost produced in a tumbling composter containing only common fruits and vegetables typical of the Costa Rican diet (Furtado et al., 2009), including both raw and cooked materials. To collect the larvae, the composter was rotated, and larvae were taken randomly. Collections were made in Heredia, province of Costa Rica (10°0’38.376’’ N & 84°7’47.964’’ W) in May 2023, where the mean temperature is 27 °C and when H. illucens larvae are found more frequently. After collection, larvae were kept in Falcon tubes and starved for eight hours to empty their ingested contents and allow the transient mycobiome to pass through. They were incubated at -80 °C for 10 min for devitalization. Disinfection was made by submerging larvae for 30 seconds in ethanol 90 %, 30 seconds in sterile distilled water, 30 seconds in sodium hypochlorite 2 %, and 30 seconds in distilled water. One millimeter of the anterior part of the larvae were cut off and the haemocoel was opened laterally using sterile scissors. The guts were carefully removed using sterile forceps and transferred individually into Eppendorf tubes containing 250 μl of 1X PBS. Samples were stored at -20 °C until their use.
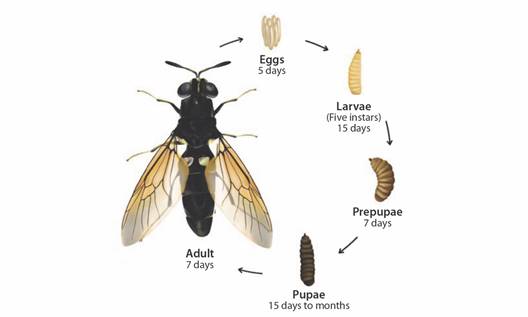
Fig. 1 Life cycle of Hermetia illucens. The development stages and average duration for each stage are illustrated (modified from De Smet et al., 2018). Only the larval stage, distinguishable by its whitish color, was used in this study.
Molecular analyses: Guts were macerated with sterile pistils, and DNA was extracted individually using a DNA isolation kit (PowerSoil, Qiagen, USA) following the manufacturer’s instructions. The eukaryotic amplicon library was generated by amplifying the ITS2 region using primers ITS3-2024F (5’-GCATCGATGAAGAACGCAGC-3’) and ITS4-2409R (5’-TCCTCCGCTTATTGATATGC-3’). Polymerase chain reaction (PCR) amplification of targeted regions was performed using specific primers connecting with barcodes. The PCR program used the following conditions: 5 min at 94 °C; 35 cycles of (30 s at 94 °C; 30 s at 50-67 °C; 30 s at 72 °C); 7 min at 72 °C. The PCR products with the proper size were selected by 1 % agarose gel electrophoresis. Each sample’s exact amount of PCR products was pooled, end-repaired, A-tailed, and further ligated with Illumina adapters. Libraries were sequenced on a paired-end Illumina platform to generate 250 bp paired-end raw reads (Illumina Novaseq, Novogene Bioinformatics Technology Co., Ltd, CA, USA).
Bioinformatic analysis: We used the DADA2 version 1.21 to process the Illumina sequenced paired-end fasta files and to generate a table of amplicon sequence variants (ASVs), which are higher resolution analogs of the traditional OTUs (Callahan et al., 2016). Briefly, we removed primers and adapters, inspected the quality profiles of the reads, filtered and trimmed sequences with a quality score < 30, estimated error rates, modeled and corrected amplicon errors, and inferred the sequence variants. Then, we merged the forward and reverse reads to obtain the full denoised sequences, removed chimeras, and constructed the ASV table. We assigned taxonomy to the ASVs with the function assignTaxonomy, of DADA2. For the fungal taxonomic assignment, we used the UNITE ITS database version 8.3, with default parameters. We carried out a second taxonomic assignment of the fungal ASVs using the tool IDTAXA of DECIPHER with the same version of UNITE as a reference and a confidence threshold > 60 %, and additionally, a third classification using the Classifier tool (Wang et al., 2007) implemented in the Ribosomal Database Project using as reference the Warcup Fungal ITS trainset V2 database (Deshpande et al., 2016). We verified and manually curated the consistency between the taxonomic assignments of the different programs. In cases of discrepancies, a comparison with the BLAST tool of NCBI was applied (Percent identity > 98 %). Sequence data were deposited at the NCBI Sequence Read Archive under accession number PRJNA1048874. Statistical analyses and visualization of results were performed with the R statistical program (R Core Team, 2023) and the Rstudio interface. Package Vegan v2.6-4 (Oksanen et al., 2022) was used to calculate alpha diversity estimators. The non-metric multidimensional scaling analyses (NMDS) weas generated using the ASV dataset, normalized into relative abundances and then converted into a Bray-Curtis similarity matrix.
Results
After the removal of low-quality reads, a total of 210 916 sequences were obtained from the samples. The average number of sequences per sample was 42 183 (ranging from 21 798 to 59 107). The fungal community was composed of 329 ASVs, according to the analysis of sequences of the ITS region. A total of 8 phyla, 21 classes, 40 orders, 67 families and 117 genera were identified. Ascomycota was the most abundant phylum with 192 561 of the sequences and 77 % of the ASVs while Glomeromycota corresponded to 15 305 of the sequences and 3 % of the ASVs. Other less abundant phyla were Rozellomycota, Basidiomycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, and Mucoromycota (Supplemental Material Table 1).
The mycobiome in the guts of H. illucens exhibited a relatively similar composition across samples (Fig. 2). The order with the highest relative abundance was Saccharomycetales, reaching 92.4 % in sample M1JL and 76.2 % in sample M5JL. Archaeosporales was the second most abundant order in four of the samples, with relative abundance ranging from 28.6 % to 10.5 %. In sample M5JL, however, Archaeosporales was not prominent; instead, Rozellida was the second most abundant order at 6.8 %. Notably, sample M5JL displayed higher diversity, with 9.1 % of sequences corresponding to 38 orders with low abundance (SMT1). Additionally, sample M5JL showed a higher presence of Erysiphales and Capnodiales, with relative abundances of 4 and 2.5 %, respectively. The most abundant genera of Saccharomycetales were Pichia, Galactomyces, and Candida, while Archaeospora and Paramicrosporidium were the most abundant genera of Archaeosporales and Rozellida respectively.
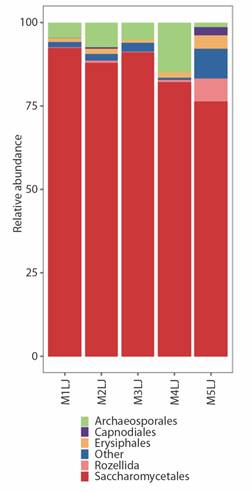
Fig. 2 Relative abundance of the fungal communities in the guts of H. illucens larvae at order level during an urban composting process.
The heat map (Fig. 3) illustrates variations in the relative abundances of the most abundant fungal genera in the sample. Pichia, the dominant genus within the Saccharomycetales, has a mean relative abundance of 65 %, ranging from 75.3 % (M3LJ) to 47.8 % (M1JL). In samples M1JL to M4JL, Galactomyces shows relative abundances ranging from 18.2 to 7.0 %, Candida from 8.4 to 8.8 %, and Archaeospora from 14.8 to 4.7 %. These three genera have low relative abundances in sample M5JL, where other genera such as Paramicrospodium (6.8 %) and Blumeria (4.0 %) are more predominant.
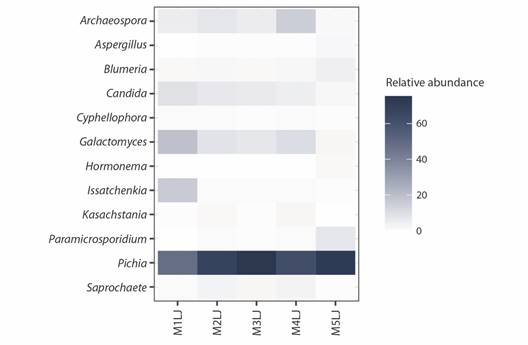
Fig. 3 Relative abundance of fungi associated with H. illucens larvae at genus level (more abundant genera are shown).
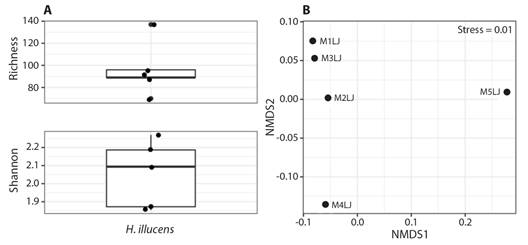
Regarding the alpha diversity indices, the sample with the highest species richness presented 137 ASVs, while the sample with the lowest species richness presented 70 ASVs. The highest average value of the Shannon Index was 2.27 and the lowest was 1.85 (Fig. 4A). However, the composition of fungal communities associated with all of the samples presented values close to 0.0 in the NDMS analysis, indicating they are relatively similar (Fig. 4B).
Discussion
In this study, we found that yeasts, in particular the genus Pichia, dominate the mycobiome of the intestinal tract of H. illucens larvae during urban composting. Recently some studies have analyzed fungal communities on BSF, most of them using colonies of larvae that were reared under laboratory conditions with different diets (Boccazzi et al., 2017; Klüber et al. 2022a; Shokry et al., 2023; Tanga et al. 2021). To our knowledge, only one study has analyzed the fungal composition of naturally occurring BSF larvae from household composts (Vitenberg & Opatovsky, 2022). All of these studies were made in regions with subtropical or temperate conditions, therefore, the present study provides a first insight into the fungal communities of naturally occurring BFS larvae in a tropical region.
The dominance of Pichia found in this investigation slightly differs with the previous study of BFS larvae reared on household compost by Vitenberg & Opatovsky (2022), where the most abundant genus was Candida (up to 70 % of the sequences), while Pichia was not common (less than 10 % of the sequences). However, Pichia has been reported as a common genus in other studies using different substrates, suggesting a stable association with BFS larval gut. Boccazzi et al. (2017), using next-generation sequencing, reported Pichia as the most abundant genus in larvae fed on vegetable waste, comprising 53.2 to 93.7 % of the sequences. Similarly, Tanga et al. (2021) observed that Pichia was the only genus that was not substrate specific, as it prevailed as an abundant group on different substrates (brewers’ spent grains, kitchen food waste, poultry manure, and rabbit manure), and reaching up to 70 % of the sequences when reared on brewers’ substrate. Klüber et al. (2022a) isolated Pichia from larvae reared on lignocellulose-rich substrates, such as palm kernel meal. In addition, Shokry et al. (2023) isolated it from larvae grown on substrates made of chicken feed, waste from fish farms, chicken nonedible parts, and kitchen waste.
Insect-yeast relations have been reported in different insects of Hymenoptera, Coleoptera, and Diptera, where H. illucens belongs to (Malassigné et al., 2021). The acquisition of yeast could be from the environment (Morales-Rodríguez et al., 2021; Rassati et al., 2019) but it could also be vertically transmitted from adults to larvae (Sacchi et al., 2008). This was recently demonstrated on Drosophila flies where yeasts acquired at the larval stage maintained through metamorphosis, and adult life, and were transmitted to offspring (Guilhot et al., 2023). The benefits obtained by insects from these relations are varied. One of the most important is nutritional, as yeast can provide insects with digestive enzymes, essential amino acids, vitamins, and sterols (Mendes et al., 2012; Stefanini, 2018). It has also been suggested that yeasts could play a role in the removal of potentially toxic compounds present in the insect’s diet (Mendes et al., 2012), as well as protection against potential pathogens for the host (Biedermann & Vega, 2020). The benefit obtained by the yeasts is less understood, but it has been related to transportation to new habitats and a stable food source (Madden et al., 2018).
Members of Saccharomycetales as Pichia and Candida have been detected in other Diptera larvae, like in some Drosophila species (Chandler et al., 2012; Hamby et al., 2012) and Aedes aegypti midguts and ovaries (Gusmão et al., 2010). They could be playing a protection role against pathogens, as the yeast of this genus is known to generate antimicrobial compounds that can inhibit the growth of some bacteria (Bajaj et al., 2013; Chelliah et al., 2016; França et al., 2015). Several strains of Pichia can produce mycocins that alter homeostasis in other sensible yeast and disrupt cell membrane function resulting in cell death (Golubev, 2006; Santos et al., 2009). The presence of Pichia and Candida in H. illucens gut might be also beneficial for its growth and development as it has been found that supplementation with Pichia increases the survival rate and decreases development time in Drosophila suzukii (Meshrif et al., 2016). In H. illucens, supplementation with Candida has proved to increase larval body weight and significantly enhanced tyrosine, purine, histidine, and vitamin B6 metabolism (Kannan et al., 2023).
The high abundance of Pichia can also have benefits boosting organic matter degradation. Inoculating compost with Pichia has been shown to accelerate the composting process as it degrades organic acids, this increases pH and allows mesophilic and thermophilic bacteria to proliferate faster, which in turn contributes to a vigorous organic matter degradation (Nakasaki et al., 2013). The capacity to tolerate low pH levels might be also related to its high dominance in this larvae gut. BSF gut presents regions with different pH values, the posterior midgut is alkaline, but the anterior region has an acidic luminal content, and the middle midgut presents pH levels around two (Tettamanti et al., 2022). Survival and efficient growth on these pH levels have been already demonstrated in this yeast (Fletcher et al., 2015; Park et al., 2018). Further research needs to be done to analyze how the location in the digestive tract and its pH influences the dominance of Pichia.
Isolation of yeasts from H. illucens larvae could be of special interest due to the conditions they could be exposed to during the composting process, as temperatures during the thermophilic phase can reach up to 70 °C (Nozhevnikova et al., 2019). It is known that thermotolerant yeasts can increase the productivity of industrial processes such as ethanol production, by eliminating the energy used to lower the temperature needed to maintain microorganisms used in fermentation (Abdel-Banat et al., 2010). Some strains of Pichia have been already tested for this purpose. They exhibited growth and ethanol production capability at temperatures up to 45 °C (Chamnipa et al., 2018; Pongcharoen et al., 2018). Pichia and Candida are useful in the production of other industrially important compounds such as malic acid, butyric acid, xylitol, biosurfactants, lipids, and enzymes among others (Kieliszek et al., 2017; Queiroz et al., 2023; Shrivastava et al., 2023). The production of these compounds can also be beneficial by utilizing thermotolerant microorganisms (Kumar et al., 2015; Mehetre et al., 2019).
In the present study, we also identified two groups that have not been reported as abundant groups in BFS larvae before. Galactomyces, another member of Saccharomycetales, have been reported on Culex pipiens and Culex theileri larvae, another insect that belongs to the order Diptera (Steyn et al., 2016). The second group is Archaespora of the order Archaeosporales, which is an arbuscular mycorrhizal (AM) fungus. Insects are known to have mutualistic relations with AM, as they could be a good source of nutrients for the insects and beneficial for the fungus due to spores being vectored by insects (Willis et al., 2013). There are no previous reports of this AM genus on insects’ guts to our knowledge. If Galactomyces and Archaespora are adapted to insects’ guts needs to be elucidated. Alternately, these species of fungus could have been common in the BFS growth environment, and therefore, highly abundant in the insect gut.
Galactomyces has biotechnological potential related to emerging pollutant removal. Chaijak et al. (2018) reported ligninolytic enzymes and phenol removal activity that can be used to treat industrial wastewater. It can also be effective in removing industrial dyes such as Methylene Blue and Azo dyes that are extensively used in textile, paper, food, and pharmaceutical industries (Contreras et al., 2019; Guo et al., 2019). Zhang et al. (2015) demonstrated the effectivity of this yeast in decomposing lincomycin, an antibiotic with a very stable structure. Isolation of these yeasts found in BFS larvae is needed for further research on their temperature tolerance range, enzymatic activities, and biotechnology applications. Additionally, while we focused on the most abundant genera found, less abundant genera could be cultivable and possess valuable properties for biotechnological applications.
To conclude, this study offers insight into the mycobiome of naturally occurring BSF larvae in a tropical region. The abundance of yeast like Pichia and Candida is consistent with previous studies, and the high dominance of Pichia in this and previous studies, suggests a stable insect-yeast relation (Boccazzi et al., 2017; Klüber et al., 2022a; Shokry et al., 2023; Tanga et al., 2021). The description of previously unreported fungal groups highlights the importance of continuing to explore the mycobiome dynamics of this larva in different habitats. The determination of dominant fungal groups can contribute to better understand the efficiency of H. illucens larva compost production, and with further reseach it may improve its use in urban composting and enhance organic waste management in these environments. Lastly, we emphasize the need for further investigation of yeast strains and other fungi groups present in this larva, as they could be adapted to more extreme conditions characteristic of this larvae’s natural habitat.
Ethical statement: the authors declare that they all agree with this publication and made significant contributions; that there is no conflict of interest of any kind; and that we followed all pertinent ethical and legal procedures and requirements. All financial sources are fully and clearly stated in the acknowledgments section. A signed document has been filed in the journal archives.











 uBio
uBio