Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.61 n.1 San José Mar. 2013
Genealogical approaches to the temporal origins of the Central American gap: Speciation and divergence in Pacific Chthamalus (Sessilia: Chthamalidae)
*Dirección para correspondencia:
Abstract
A large section of the tropical Eastern Pacific coastline is nearly devoid of reef or consolidated habitat, and is known as the Central American Gap as it is associated with a biogeographic transition in fish and invertebrate species. We analyze phylogeographic data for intertidal barnacles (Chthamalus) to identify relevant temporal patterns that describe the origins of this biogeographic transition (the Mexican-Panamic Transition Zone). These contrasts of populations on either side of the transition zone include two pairs of closely related species (C. panamensis and C. hedgecocki; C. southwardorum and a Southern form of C. southwardorum), as well as gene flow data within one species (C. panamensis) that currently is found on both sides of the boundary between provinces. Using sequence data from a prior phylogenetic study, we used traditional (net nucleotide divergence) measures as well as coalescent analyses that incorporate the isolation-migration model to identify the likely time of separation between Northern and Southern taxa in two species pairs. A total of 67 individuals were sequenced at two mitochondrial (cytochrome c oxidase I, 16S) and one nuclear (elongation factor 1-alpha) gene regions. Our analyses indicate that the regional isolation of these intertidal barnacles occurred approximately 315-400kya, with subsequent expansion of C. panamensis from the Southern region into the North much more recently. There are insufficient survey data to conclusively document the absence of species from this group within the Central American Gap region near the Gulf of Tehuantepec. However, appropriate habitat is quite sparse in this region and other environmental factors, including upwelling and water temperature, are likely to be associated with isolation of many species in the Mexican and Panamic provinces sensu stricto. Some taxa may maintain gene flow across this region, but very few genetic studies have been completed on such taxa. Until further work is done, distinguishing between prior hypotheses of a faunal gap, or a faunal transition zone, is somewhat speculative. Additional taxonomic revision will be necessary in Chthamalus but is beyond the scope of this paper.
Key words: Pacific, intertidal, barnacle, phylogeography, transition zone, Central American Gap, Chthamalus.
Resumen
La taxonomía del complejo de especies de cirripedios (Chthamalus) se ha confundido en la literatura desde hace casi 30 años, por lo tanto analizamos datos de su filogeografía para identificar modelos temporales relevantes que describan los orígenes de la zona de transición entre las provincias Mexicana y Panameña. Estos contrastes de poblaciones a ambos lados de la zona de transición incluyen a dos pares de especies estrechamente relacionadas, así como datos de flujo de genes dentro de una especie que actualmente es encontrada en ambos lados del límite entre provincias. Usando datos de secuencia de un estudio previo de filogenética, usamos medidas tradicionales, así como análisis de coalescencia que incorporan el modelo de migración y aislamiento para identificar el tiempo probable de la separación entre los taxa del norte y del sur en dos pares de especies. Nuestros análisis indican que el aislamiento regional de estos ciripedios ocurrió aproximadamente hace 315-400 mil años, con una extensión subsecuente de Chthamalus panamensis de la región del sur hacia el norte mucho más reciente. No hay datos suficientes para documentar conclusivamente la ausencia de especies de este grupo dentro de la región de Centro América cerca del Golfo de Tehuantepec. Sin embargo, el hábitat apropiado es bastante escaso en esta región y otros factores ambientales, incluyendo corrientes y temperatura acuática, probablemente están relacionados con el aislamiento de muchas especies en estas provincias. Algunos taxa pueden mantener el flujo de genes a través de esta región, pero muy pocos estudios genéticos han sido realizados en tales taxa. Hasta que no se desarrollen trabajos adicionales, la distinción entre hipótesis previas de un “gap” faunal o de una zona de transición faunal es algo especulativo.
Palabras clave: océano Pacífico, zona intermareal, cirripedios, filogeografía, zona de transición, Centro América.
The conundrum of marine speciation has become a common subject of investigation in recent years. The long dispersal potential of many marine organisms and the paucity of absolute physical barriers would suggest the existence of a small number of widespread species, and yet in actuality marine systems are incredibly diverse, with high levels of endemism (Palumbi 1994). What we have come to recognize is the dynamic nature of habitat and species distributions through time. Histories of allopatry, expansion, and habitat loss are not always readily apparent from contemporary species distributions (McGovern et al. 2010). It is of interest to characterize these historical events when they appear to have influenced large numbers of taxa. In particular, what ancient and contemporary forces separate particular marine provinces, and ecoregions within provinces (Spalding et al. 2007)? The origins of biogeographic transition zones may be environmental in nature (associated with habitat) or may be caused by ancient events that are no longer concordant with environmental patterns.
One such transition zone is found along the Mexican coast of the Tropical Eastern Pacific (TEP) ocean between roughly 13-15°N latitude. Called the “Central American Gap” (CAG) by some (Hastings 2000, Pitombo & Burton 2007) and the Mexican-Panamic transition zone (MPTZ) in other sources (Laguna1990), this region separates the Mexican and Panamic sensu stricto marine provinces with a “stretch of coastline over 1 200km in length dominated by sandy/muddy shores and mangrove- lined lagoons” (Hastings 2000). This region is known to influence the distribution of rocky shore and reef fish, as well as invertebrates, and is a likely factor in the separation of cryptic species in the region.
The widespread barnacle genus Chthamalus has been the subject of several biogeographic studies in the TEP (Laguna 1990, Wares 2001, Pitombo & Burton 2007, Wares et al. 2009). Very low levels of morphological variation have made determining species distributions difficult, and until recently many species in the ‘fissus group’ (defined by two minute but verified synapomorphies) were considered a single species or species complex (Dando 1987). Recent phylogenetic studies have indicated that cryptic diversity is yet to be fully explored in this group, with fine-scale morphological variation and DNA barcoding being used to fully describe two new species, C. hedgecocki and C. southwardorum (Pitombo& Burton 2007). Based on morphology alone, the latter is reported to be distributed from Bahio Kino, Mexico, to Puerto Chicama, Perú, but molecular data were previously only available from the Northernmost samples. Pitombo & Burton (2007) predicted that a sibling species group may still exist in this widespread species range, which crosses the CAG; this prediction was founded in part on allozyme data presented by Dando & Southward (1980), but no formal revision or analysis has been completed to date.
To evaluate both the potential for further cryptic diversity and the insights this diversity can provide for understanding the temporal habitat dynamics of the CAG, we have analyzed previously sampled populations of C. southwardorum from sites North and South of this faunal transition and use mitochondrial and nuclear sequence-based markers to measure genetic differentiation among these sites. The same methods are applied to a closely related species pair, C. hedgecocki and C. panamensis, that are found in more wave-exposed coastal habitats. We use analytical approaches based on coalescent theory that allow the co-estimation of divergence times among populations and the migration rates among them (the isolation-migration model; Hey & Nielsen 2004). This approach enables us to separate the effects of ancestral polymorphism and gene flow at loci where alleles are shared between the two regions. Using substitution rates generated from a phylogenetic analysis of the genus (Wares et al. 2009), we have also evaluated the likely time frame at which these regional populations could have become isolated from one another.
Materials and Methods
Study system: The taxonomy of this species complex has been confused in the literature for nearly 30 years. This has been in part because although data were available to indicate there were two pairs of Chthamalus along this coast (Northern and Southern pairs of wave-exposed habitat and wave-sheltered habitat species), the taxonomy was only partially updated by Pitombo & Burton (2007), after a series of intervening papers that applied unofficial and inconsistent nomenclature. The four species evaluated here, with an attempt at taxonomic history for each, are: C. panamensis Pilsbry – though its exact distribution has been in question given the difficulty in field identification of this species, panamensis has been consistently used for this taxon. Found in wave-exposed coastal habitats, this species resides primarily in Southern locations in the TEP. Pitombo & Burton (2007) indicated that it may range as far North as 20°N.
C. hedgecocki Pitombo & Burton – originally part of the C. fissus (Darwin) species complex (Hedgecock 1979), this species was distinguished from the rest of the complex using phenotypic and electrophoretic data by Dando & Southward (1980) and unofficially called C. “mexicanus”. The same name was also used by Laguna (1990), but this species was officially described as C. hedgecocki in Pitombo & Burton (2007). It is found in waveexposed coastal habitats, primarily at Northern locations in this region.
C. southwardorum Pitombo & Burton – as with C. hedgecocki, this was originally distinguished as a separate taxon by Dando & Southward (1980) and unofficially called C. “cortesianus” (Laguna 1990). Formally described as C. southwardorum in Pitombo & Burton (2007), it can be found in wavesheltered coastal habitats, solely at Northern locations in this region.
C. southwardorum “Farfan” form – named for the putative type locality in Panama (at which 100% of individuals were of this form in 1978, P. Dando pers. comm.), populations of this form were identified as distinct by Dando & Southward (1980) but with no formal description. It has been confused in the literature with C. southwardorum sensu stricto, which may influence our understanding of regional biogeography. The “Farfan” form is found in wave-sheltered coastal habitats, solely at Southern locations in this region. Recent data further suggest a difference between the forms (Wares et al. 2009), though the nomenclature of the C. southwardorum A and B clades is inconsistent in that source.
Note that other congeneric species, including C. fissus and C. anisopoma, may be sympatric with these taxa in the Gulf of California or along the Baja California peninsula, but are more readily distinguished from this complex.
Chthamalus individuals were collected in 2005-2006 from seven primary locations in the TEP (Fig. 1, Table 1) as part of a global phylogeny of the group (Wares et al. 2009). Species in this complex are largely indistinguishable in the field based on external morphology (Pitombo & Burton 2007; P. Dando, pers. comm.). Thus, it was not possible to sample directly for particular species, but rather the available specimens were defined using standard DNA barcoding approaches with the mitochondrial cytochrome oxidase I (COI) locus (Wares et al. 2009). Specimens of C. southwardorum were found in five locations (Table 1, Fig. 1), with three locations North of the CAG and two locations to the South. Specimens of C. hedgecocki and C. panamensis were identified from three additional locations (Table 1). Molecular PCR and sequencing techniques are described in Wares et al. (2009). Two sequences at the 16S locus from the previous study, one each from C. panamensis and C. southwardorum, were excluded from analysis after being identified as either erroneously labeled or product of contamination. Removal of these two sequences results in the reciprocal monophyly of C. southwardorum and the clade containing C. hedgecocki and C. panamensis (Wares et al. 2009).
Phylogenetic analysis: Here we use previously reported (Wares et al. 2009) aligned sequence data for two mitochondrial gene regions (COI and 16S rDNA) and one nuclear locus (elongation factor 1-alpha, EF1) from the locations indicated in table 1 and figure 1. Additional nuclear sequence data are available for these specimens, but provide little phylogenetic signal at this level and considerable homoplasy (Wares et al. 2009). The separation of locations by the CAG is used for all a priori structuring of data in subsequent analysis. Due to difficulty in amplifying some loci from some individuals, not all locations/individuals/loci are equally represented (Table 1).
Sequence data at the EF1 nuclear locus were directly sequenced and polymorphism within sequences was recoded as ambiguity, either unknown “N” in mitochondrial data or appropriate IUPAC ambiguity codes for nuclear data. Phylogenetic reconstructions were performed on the concatenated data using MrBayes v3.1.2 (Ronquist & Huelsenbeck 2003) with model estimation unlinked across loci. Bayesian estimation followed 5x106 generations of four MCMC chains, with a burn-in fraction of 25%. Data from the EF1 locus were analyzed with ambiguity represented as heterozygous sites for phylogenetic analysis.
Coalescent analysis: We analyzed the data as multilocus observations from the Northern and Southern sites using the program IMa2 (Hey & Nielsen 2004), which applies the isolation-migration model for estimation of the relative importance of migration and time since divergence. The IMa2 program uses a Markov Chain Monte Carlo method for assessing posterior probability densities of six model parameters: time of divergence, migration from North to South, migration from South to North, and effective population sizes for the two regional populations as well as their ancestral source population. Substitution rate estimates for each locus (Wares et al. 2009) were included to allow the time since divergence to be estimated from the coalescent model. These rates, in units of substitutions per locus/year, are: mtCOI (1.83x10-5); mt16S (7.65x10-6); nEF1 (1.86x10-6). The nEF1 data were separated into haplotypes for coalescent analysis using the PHASE algorithm (Stephens et al. 2001) as implemented in DNAsp (Librado & Rozas 2009). This analysis was repeated for the taxon pair C. hedgecocki – C. panamensis, and for Northern and Southern populations of C. panamensis. Priors allowed for population mutation rates ranging from 10-100 (most likely estimate based on π of 15 was used for final calculations with a maximal range of 30, see below), migration parameter from 0-5, and separation time of four coalescent units. A burn-in period of 10 000 steps was used, followed by a run duration of 108 steps with 20 geometrically heated chains.
Given the potential for spatial and temporal concordance of separation between the Northern and Southern species pairs, temporal association was tested using the msBayes pipeline (Hickerson et al. 2006). This approach uses approximate Bayesian computation (ABC) simulation approaches to evaluate the most likely isolation scenarios associated with empirical data; in our case we used 106 simulated data sets and an upper prior on ancestral effective size equal to the upper prior on current effective size (otherwise default parameters for analysis).
Results
Phylogenetic analysis: Data from C. southwardorum, C. panamensis, and C. hedgecocki used in this study are as in Wares et al. (2009) but with more focused analysis of genealogical relationships. Genbank voucher numbers for all sequence data are provided in Wares et al. (2009). Bayesian phylogenies for each taxon pair are shown in figures 2 and 3; similar results were obtained from likelihood bootstrap analysis and parsimony analysis (results not shown). Net nucleotide divergence (Nei & Li 1979)Li - Proceedings of the National Academy of Sciences - 1979 between the Northern and Southern populations of C. southwardorum was 0.0288±0.0043 for COI, 0.0047±0.0019 for 16S, and 0.0019±0.0013 for EF1 (0.0044±0.0016 after haplotype phase calculated). Given the substitution rates indicated above, these dA values would suggest divergence times of approximately 5.11x105, 1.69x105, and 5.32x105 years, respectively (mean 4.04x105).
For comparison, the divergence of the C. panamensis – C. hedgecocki species pair involves net nucleotide divergences of 0.0210±0.0040 for COI, 0.0091±0.0051 for 16S, and 0.0034±0.0025 for EF1. These values suggest divergence times at each locus of approximately 3.73x105, 3.27x105, 4.81x105 (mean 3.98x105) years.
Coalescent analysis: Homogeneity of populations in the Northern and Southern regions for each taxon was indicated from Hudson’s (2000) Snn statistic (p>0.05 in all cases). Coalescent analysis of the nEF1 locus produced similar results for C. southwardorum whether using phased or unphased sequences (results not shown); each supported negligible migration between populations of C. southwardorum, though the age estimation based on the nEF1 locus alone is relatively uninformative as there is considerably less polymorphism. Overall, the IMa2 analyses ran sufficiently long to reach good convergence as determined by effective sample size (ESS) values and very similar likelihood distributions (Fig. 4). Effective size estimates were consistent between runs, with the ancestral effective population size 9.27-9.54x104, and current effective population sizes of 1.45-1.47x105 (Northern) and 2.36x105 (Southern). Migration rate estimates between the two regional populations were consistently negligible (≤0.0001 migrants per 1 000 generations in either direction). Posterior probability distributions of divergence time estimates peaked approximately 340kya in all runs, with minimum 95% limits at around 220kya (Fig. 4). High 95% limits are not reported because the distributions have fat tails and thus these maximum (and mean) values are conditioned on the prior. Our time distribution range was set to correspond with the time since the rise of the Isthmus of Panama (~three million years ago).
An identical analysis performed on the C. panamensis – C. hedgecocki species pair was consistent with the above results. Migration rate estimates between the two taxa were consistently negligible (≤0.001 migrants per 1 000 generations in either direction). However, the posterior probability distribution of divergence time estimates exhibited an increasing function with time, meaning no useful estimate of divergence is possible with these data.
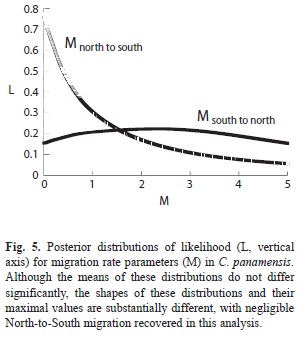
Full analysis of the C. panamensis data in IMa2 indicated maximal population sizes in current Northern and Southern regions (e.g. limited only by the prior, as discussed here prior maximum on theta is 30; see Discussion). The ancestral population substitution rate estimate was approximately what would be predicted from π (13.9; 95% range 8.26- 24.38). Migration rate 2Nm from the South to the North was estimated at (mean) 2.46 (0.16, 4.84) and from North to South at 1.48 (0.03, 4.58). Plots of migration rates between the two regions are substantially different, with a maximal posterior probability from South to North of 2.36 and from North to South of 0.003 (Fig. 5). The time of separation of the Northern and Southern populations, coestimated with migration, is approximately 166 830 years given our substitution rate estimates (95% range of 47 400-922 000).
Analysis in msBayes of the coalescentbased divergence times of the two species pairs indicated high probability that separation of the two lineages of C. southwardorum, and C. panamensis and C. hedgecocki, was temporally concordant. With local multinomial logit regression, the posterior probability for a single divergence time between the two pairs was 0.929, with remaining probability allowed for two distinct divergence times. The estimated time of simultaneous divergence using this approach, with the calculations of Huang et al. (2011), is 315 000 years (assuming a generation time of one year and the substitution rates noted above).
Discussion
Our results suggest a temporal pattern of increasing isolation among TEP chthamalid barnacles; the deepest separation of the four species evaluated suggests that the sheltered-exposed species pairs were separated approximately one million years ago, based on net nucleotide divergences ranging from 7.5- 8% at mitochondrial COI. The focus of this study, the potential for geographic isolation of species across the CAG, allows us to evaluate the potential for a single environmental change promoting separation of Northern and Southern lineages of both the wave-sheltered and waveexposed forms. Based on classical approaches such as net nucleotide divergence, as well as two different coalescent estimators, our results are surprisingly consistent in estimating a separation time between C. hedgecocki and C. panamensis, and the type and “Farfan” forms of C. southwardorum, of approximately 400kya, corresponding with the timing of one of the mid-Pleistocene high sea level stands (Muhs et al. 2004). These results also indicate strongly (92.9% posterior probability) that isolation of C. southwardorum Northern and Southern populations was temporally concordant with the isolation of C. hedgecocki and C. panamensis. It remains to be determined if this apparent vicariant event is associated with a biogeographic transition zone between the Mexican and Panamic provinces (requiring further analysis of species that do cross the region) or a true habitat and faunal gap (Audzijonyte & Vrijenhoek 2010).
Estimation of gene flow between these two taxon pairs (treating the Southern populations of C. southwardorum as a distinct taxon, or at least separate population) indicates negligible gene flow between the Northern and Southern forms of both the wave-sheltered (C. southwardorum) and wave-exposed (C. hedgecocki, C. panamensis) taxa. However, additional analysis of the one taxon for which we have samples spanning the region, C. panamensis, indicates that a more recent (~160kya) expansion from the Southern populations to the North is likely to be the cause of the current distribution, both geographic and in terms of genetic diversity. All of these results are also supported by the inclusion of data from one additional nuclear locus, though sample sizes were too small to provide sufficient additional information (results not shown).
Conclusions from our analyses must be tempered by the sparse samples available for this study. Most of the sequence data are from a single mitochondrial locus, sequenced at two distinct regions (mtCOI and mt16S). It should be emphasized that the data obtained were initially part of a separate phylogenetic analysis in which cryptic species were distinguished, without reference to external morphology in field collections. Samples of C. southwardorum were not recovered from sites immediately North of the CAG, nor were any chthamalids identified within the CAG during the NSF-funded Tropical Eastern Pacific Expedition of Newman in 1978 (W. Newman, P. Dando, E. Southward, pers. comm.). Thus, the opportunistic sample size reflects both the sampling effort and PCR success in these samples. Considering the reciprocal monophyly of regional samples at mitochondrial gene regions and extensive lineage sorting at nuclear gene regions, the results appear to be robust to sample size (Rosenberg 2007). One problematic result of the coalescent analyses was the estimate of Ne in populations of C. panamensis being predicated on the maximum of the prior distribution. This is often the case when the data are limiting, and may be associated with the signal of expansion noted above.
Briggs (1974) and Brusca & Wallerstein (1979) first indicated that a zoogeographic break in the marine fauna existed at approximately 15° N (roughly the Gulf of Tehuantepec). There are a number of strong environmental shifts associated with this coastal region (Udvardy 1975, Hayden & Dolan 1976). There are significant sea surface temperature and wind stresses associated with the Gulf of Tehuantepec (Sun & Yu 2006), generating strong upwelling in the Gulf and offshore movement of surface waters (Kessler 2006, Barton et al. 2009). Upwelling patterns alone have previously been associated with divergence of regional barnacle populations (Dando & Southward 1981). The coast in this region is predominantly sandy (Robertson & Cramer 2009, W. A. Newman, pers. comm.), with little consolidated habitat for barnacles or reef organisms to persist. A remaining concern is whether this region represents a true faunal ‘gap’ or transition zone, a question that requires integration across more taxa (Audzijonyte & Vrijenhoek 2010).
While certain groups of species that require consolidated habitat have reduced diversity or abundance in this region, indicating a gap, the patterns of distribution in most taxa may still be consistent with an overlap zone between two biogeographic provinces. Reid (2002) provides such data for littorinid snails, and Laguna (1990) illustrates this in the distribution of barnacles along the tropical Eastern Pacific coast. Notably, there is a clear demarcation between the Mexican and Panamic provinces sensu stricto, but there are a number of barnacle species defined by Laguna (1990) as being of the Panamic province sensu lato that do broadly overlap this transition zone and the CAG.
The present study makes it apparent that at least one of these species (called C. “mexicanus” at the time, but presently C. southwardorum) does not readily cross this region. Though the samples in the present study are sparse, it is consistent with information from the TEPE survey, with no representatives of C. southwardorum (or the “Farfan” form) collected between 9°54’ N and 21°30’ N. Based on allozyme data from the earlier survey, what is now C. southwardorum was found in high abundance in the Northern Gulf of California (and 100% of our Chthamalus from Bahia Kino are C. southwardorum), but collections between Bahia Chamela (19°30’ N), Salina Cruz (16° N) and at Isla Sacrificios (15°42’ N) recovered only C. hedgecocki (P. Dando, W. Newman, pers. comm.). In the Southern range, as far North as Murciélagos, Costa Rica (10°48’ N) only C. panamensis were recovered at the time. Certainly, even when multiple species are present, one may be extremely rare or microhabitat specific. It is an open question whether genetic analysis of other species in this region (group IV in Laguna 1990) would indicate a population genetic pattern consistent with the divergence of C. southwardorum.
The generality of results presented here to the broader faunal distribution patterns of the Tropical Eastern Pacific remains ambiguous. It is clear that for some taxa that require (or form) consolidated substrate or reef habitat, the lack of such habitat in this region (the Central American Gap) is a significant demarcation in diversity along this coast (Hastings 2000, Díaz- Jaimes et al. 2006, Vargas et al. 2008). Studies that have focused only on the apparent distribution of species suggest, however, that the effect of these habitat gaps on marine diversity is minimal and the association of transition zones with gaps like the CAG is possibly stochastic (Robertson & Cramer 2009). Robertson & Cramer (2009) note that because the appearance of the CAG as a “distinctive entity” in their analysis is in part because of an absence of data, rather than information on the potential for migration or gene flow across the region, additional range-wide genetic analysis of fish and other taxa is warranted. The small number of population genetic studies that span this region suggest there are many cryptic populations along the TEP coast (Hellberg 1998, Arnaud et al. 2000, Díaz-Jaimes et al. 2006, Wares et al. 2009, Saarman et al. 2010), and in many cases the break in diversity could be spatially associated with the CAG (Díaz-Jaimes et al. 2006, Hurtado et al. 2007, Pitombo & Burton 2007). In the intertidal snail Nerita, two species are known to span this region; one exhibits a strong break while the other does not (Hurtado et al. 2007) L.A. Frey, M . Gaube , P. Pfeiler , E . Markow, T.A. Geographical subdivision, demographic history and gene flow in two sympatric species of intertidal snails, Nerita scabricosta and Neritafuniculata ,from the tropical eastern Pacific MarineBiology Marine Biology Mar.Biol. 1863-1873 151 5 Baja California differentiation fish gulf-of-california human mitochondrial-NA patterns penaeus-stylirostris population growth rockyshore sequences 2007 0025-3162 Doi10.1007/S00227-007-0620-5, suggesting that knowledge of both habitat and interactions between life history, behavior, and physical oceanography may be necessary to predict the effect of this habitat break on coastal diversity.
Robertson & Cramer (2009) noted that the CAG may have formed recently relative to the origins of many taxa in their study, and that more work must be done to assess intraspecific genetic variation among populations of TEP fish taxa on either side of the CAG. Although they argue that the distribution of species suggests gene flow has the potential to be quite high even across this substantial habitat gap – placing the emphasis on other forms of environmental variation for driving the diversity patterns of this coast – we show that for species with long (2-3 weeks; Miller et al. 1989) effective larval dispersal, gene flow is negligible. On recent time scales, the CAG may be promoting intraspecific diversification. It is important to note that the CAG may have distinct effects on different groups of organisms; the delimitation based on absence of reef fishes (Robertson & Cramer 2009) is quite narrow (15-16° N). It may be that more vagile organisms like pelagic fish are less sensitive to such barriers (Lessios & Robertson 2006), while benthic invertebrates or demersal fish have less opportunity to complete such a journey and successfully recruit (Waters et al. 2007). For intertidal invertebrates, and in particular the Cirripedia, this region is certainly an area of faunal transition (Laguna 1990), separating the Mexican and Panamic provinces. The mechanism for diversification may be more than simply habitat availability, as Laguna (1990; also W. A. Newman, P. Dando, pers. comm.) points to earlier work that suggests local currents and upwelling could act as dispersal barriers in this depositional shoreline region. Eventually, with addition of more locational information and microhabitat environmental data, there may be more explicit ways to evaluate the mechanism by which these species have been separated (Glor & Warren 2011). A more thorough spatial survey will be necessary to fully generate models that are useful for microhabitat identification (Lozier et al. 2009).
Assuming an average age of the isolating event between these Northern and Southern lineages of around 400kya (Fig. 4), it is not clear what model of isolation best fits these data. The analytical approaches used here are necessarily indirect (Slatkin 1985) and can have their limitations (Wares & Cunningham 2005), but the concordant results between two distinct estimators suggests our temporal estimate is robust. Hickerson & Meyer (2008) developed a method for comparing the likelihood of “soft” vicariance (in which an effectively panmictic species then becomes isolated across some stretch of its geographic range because of habitat or climate change) versus peripatric events in which a colonization from one side of the boundary establishes a new lineage on the other side. However, these methods require data from a larger number of taxa. Separating the influences of temporal isolation and recurrent gene flow is often critical in assessing alternate historical scenarios that could produce patterns of extant diversity (McGovern et al. 2010).
The question of whether the two subgroups, Northern and Southern, of C. southwardorum merit taxonomic revision is a question that has been addressed but only informally. The regional populations were considered likely to represent separate taxa based on the relative mobilities of Ald and Pgk allozymes, and some coloration differences between exposed-shore populations and sheltered populations in the Mexican (now C. hedgecocki and C. southwardorum) and Panamic (now C. panamensis and the Southern group of C. southwardorum) provinces (Dando & Southward 1980), and tergal groove morphology separating the two Panamic species. The exposed coast forms have orange tergal-scutal tissue flaps (with two pale spots on the flap in C. hedgecocki), and the sheltered coast forms are brown or “orange-brown” for the same tissues (P. Dando, pers. comm.). Pitombo & Burton (2007) also note that C. southwardorum could consist of two sibling species, but this was untested in that study. At present it is simply referred to as the “Farfan” form to avoid any further taxonomic confusion in this group; full taxonomic designation will require integration of phenotypic and genetic identifiers, and will be handled separately. What may be of most general interest about these results – particularly in designing future studies of taxa that apparently cross this coastal region – is that for Chthamalus, the speciation of inshore (sheltered) and outer coast (exposed) forms clearly happened first (Wares et al. 2009), followed by the latitudinal divergence associated here with the CAG. The generality of this result should be examined further.
Acknowledgments
The authors would like to express their appreciation first and foremost to Bill Newman, Eve Southward, and Paul Dando, who dug into their files to recover historical surveys of the chthamalid fauna to assist our study. We certainly appreciate the decades of work these scientists and the late Alan Southward put into study of the Cirripedia, and these species in particular. Fabio Pitombo, Liza Gomez Daglio, and John Binford contributed significantly to early data collection efforts in this project. Carolyn Embach provided technical editing services. This work was partially supported by the National Science Foundation (OCE-1029526 to JPW) and the U.S.-Israel Binational Science Foundation (grant # 2004/239 to JPW).
References
Arnaud, S., M. Monteforte, N. Galtier, F. Bonhomme & F. Blanc. 2000. Population structure and genetic variability of pearl oyster Pinctada mazatlanica along Pacific coasts from Mexico to Panama. Conserv. Genet. 1: 299-307. [ Links ]
Audzijonyte, A. & R.C. Vrijenhoek. 2010. When gaps really are gaps: Statistical phylogeography of hydrothermal vent invertebrates. Evolution 64: 2369-2384. [ Links ]
Barton, E.D., M.F. Lavin & A. Trasvina. 2009. Coastal circulation and hydrography in the Gulf of Tehuantepec, Mexico, during winter. Cont. Shelf Res. 29: 485-500. [ Links ]
Briggs, J.C. 1974. Marine Zoogeography. McGraw-Hill, New York, USA. [ Links ]
Brusca, R.C. & B.R. Wallerstein. 1979. Zoogeographic patterns of idoteid isopods in the northeast Pacific, with a review of shallow-water zoogeography for the region. Bull. Biol. Soc. Wash. 3: 67-105. [ Links ]
Dando, P.R. 1987. Biochemical genetics of barnacles and their taxonomy, p. 73-88. In A.J. Southward (ed.). Barnacle Biology. A.A. Balkema, Rotterdam, The Netherlands. [ Links ]
Dando, P.R. & A.J. Southward. 1980. Speciation studies on chthamalid barnacles from the tropical Pacific Coast of North America: a combined approach using biochemical genetics, morphology and ecology. 2nd International Congress of Systematic and Evolutionary Biology Chemosystematics: Principles and Practice, Vancouver, British Columbia, Canada. [ Links ]
Dando, P.R. & A.J. Southward. 1981. Existence of Atlantic and Mediterranean Forms of Chthamalus-Montagui (Crustacea, Cirripedia) in the Western Mediterranean. Mar. Biol. Lett. 2: 239-248. [ Links ]
Díaz-Jaimes, P., M.D. Barbosa-Saldaña & M. Uribe-Alcocer. 2006. Allozyme variation in eastern Pacific brown shrimp Farfantepenaeus californiensis populations. Fish. Sci. 72: 696-698. [ Links ]
Glor, R.E. & D. Warren. 2011. Testing ecological explanations for biogeographic boundaries. Evolution 65: 673-683. [ Links ]
Hastings, P.A. 2000. Biogeography of the Tropical Eastern Pacific: distribution and phylogeny of chaenopsid fishes. Zool. J. Linn. Soc. 128: 319-335. [ Links ]
Hayden, B.P. & R. Dolan. 1976. Coastal marine fauna and marine climates of the Americas. J. Biogeogr. 3: 71-81. [ Links ]
Hedgecock, D. 1979. Biochemical genetic variation and evidence of speciation in Chthamalus barnacles of the tropical eastern Pacific ocean. Mar. Biol. 54: 207-214. [ Links ]
Hellberg, M.E. 1998. Sympatric sea shells along the sea’s shore: the geography of speciation in the marine gastropod Tegula. Evolution 52: 1311-1324. [ Links ]
Hey, J. & R. Nielsen. 2004. Multilocus Methods for Estimating Population Sizes, Migration Rates and Divergence Time, With Applications to the Divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747-760. [ Links ]
Hickerson, M.J. & C.P. Meyer. 2008. Testing comparative phylogeographic models of marine vicariance and dispersal using a hierarchical Bayesian approach. BMC Evol. Biol. 8: 322-322. [ Links ]
Hickerson, M.J., E.A. Stahl & H.A. Lessios. 2006. Test for simultaneous divergence using approximate Bayesian computation. Evolution 60: 2435-2453. [ Links ]
Huang, W., N. Takebayashi, Y. Qi & M.J. Hickerson. 2011. MTML-msBayes: Approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity. BMC Bioinformatics 12: 1. [ Links ]
Hudson, R.R. 2000. A new statistic for detecting genetic differentiation. Genetics 155: 2011-2014. [ Links ]
Hurtado, L.A., M. Frey, P. Gaube, E. Pfeiler & T.A. Markow. 2007. Geographical subdivision, demographic history and gene flow in two sympatric species of intertidal snails, Nerita scabricosta and Nerita funiculata, from the tropical eastern Pacific. Mar. Biol. 151: 1863-1873. [ Links ]
Kessler, W.S. 2006. The circulation of the eastern tropical Pacific: A review. Prog. Oceanogr. 69: 181-217. [ Links ]
Laguna, J.E. 1990. Shore Barnacles (Cirripedia, Thoracica) and a Revision of their Provincialism and Transition Zones in the Tropical Eastern Pacific. Bull. Mar. Sci. 46: 406-424. [ Links ]
Lessios, H.A. & D.R. Robertson. 2006. Crossing the impassable: genetic connections in 20 reef fishes across the eastern Pacific barrier. Proc. R. Soc. Lond., Ser. B: Biol. Sci. 273: 2201-2208. [ Links ]
Librado, P. & J. Rozas. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451-1452. [ Links ]
Lozier, J.D., P. Aniello & M.J. Hickerson. 2009. Predicting the distribution of Sasquatch in western North America: anything goes with ecological niche modelling. J. Biogeogr. 36: 1623-1627. [ Links ]
McGovern, T.M., C.C. Keever, C.A. Saski, M.W. Hart & P.B. Marko. 2010. Divergence genetics analysis reveals historical population genetic processes leading to contrasting phylogeographic patterns in codistributed species. Mol. Ecol. 19: 5043-5060. [ Links ]
Miller, K.M., S.M. Blower, D. Hedgecock, & J. Roughgarden. 1989. Comparison of larval and adult stages of Chthamalus dalli and Chthamalus fissus (Cirripedia: Thoracica). J. Crust. Biol. 9: 242-256. [ Links ]
Muhs, D.R., J.F. Wehmiller, K.R. Simmons & L.L. York. 2004. Quaternary sea level history of the United States, p. 147-183. In A.R. Gillespie, S.C. Porter & B.F. Atwater (eds.). The quaternary period in the United States. Elsevier, Amsterdam, The Netherlands. [ Links ]
Nei, M. & W.H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269-5273. [ Links ]
Palumbi, S.R. 1994. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 25: 547-572. [ Links ]
Pitombo, F.B. & R.O.N. Burton. 2007. Systematics and biogeography of Tropical Eastern Pacific Chthamalus with descriptions of two new species ( Cirripedia , Thoracica ). Zootaxa 30: 1-30. [ Links ]
Reid, D.G. 2002. The genus Nodilittorina von Martens, 1897 (Gastropoda: Littorinidae) in the eastern Pacific Ocean, with a discussion of biogeographic provinces of the rocky-shore fauna. Veliger 45: 85-170. [ Links ]
Robertson, D.R. & K.L. Cramer. 2009. Shore fishes and biogeographic subdivisions of the Tropical Eastern Pacific. Mar. Ecol. Prog. Ser. 380: 1-17. [ Links ]
Ronquist, F. & J.P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. [ Links ]
Rosenberg, N.A. 2007. Statistical tests for taxonomic distinctiveness from observations of monophyly. Evolution 61: 317-323. [ Links ]
Saarman, N.P., K.D. Louie & H. Hamilton. 2010. Genetic differentiation across eastern Pacific oceanographic barriers in the threatened seahorse Hippocampus ingens. Conserv. Genet. 11: 1989-2000. [ Links ]
Slatkin, M. 1985. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16: 393-430. [ Links ]
Spalding, M.D., H.E. Fox, G.R. Allen, N. Davidson, Z.A. Ferdaña, M. Finlayson, B.S. Halpern, M.A. Jorge, A. Lombana, S.A. Lourie, K.D. Martin, E. McManus, J. Molnar, C.A. Recchia & J. Robertson. 2007. Marine Ecoregions of the World: A Bioregionalization of Coastal and Shelf Areas. Bioscience 57: 573-573. [ Links ]
Stephens, M., N. Smith & P. Donnelly. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978-989. [ Links ]
Sun, F.P. & J.Y. Yu. 2006. Impacts of Central America gap winds on the SST annual cycle in the eastern Pacific warm pool. Geophys. Res. Lett., 33, L06710. (doi:1 0.1029/2005GL024700). [ Links ]
Udvardy, M.D.F. 1975. A classification of the biogeographical provinces of the world. IUCN Occasional Paper No. 18, Morges, Switzerland. [ Links ]
Vargas, S., H.M. Guzman & O. Breedy. 2008. Distribution patterns of the genus Pacifigorgia (Octocorallia: Gorgoniidae): track compatibility analysis and parsimony analysis of endemicity. J. Biogeogr. 35: 241-247. [ Links ]
Wares, J.P. 2001. Patterns of speciation inferred from mitochondrial DNA in North American Chthamalus (Cirripedia: Balanomorpha: Chthamaloidea). Mol. Phylogen. Evol. 18: 104-116. [ Links ]
Wares, J.P. & C.W. Cunningham. 2005. Diversification predates glaciation in Balanus glandula. Biol. Bull. 208: 60-68. [ Links ]
Wares, J.P., M.S. Pankey, F.B. Pitombo, L.G. Daglio & Y. Achituv. 2009. A “shallow phylogeny” of shallow barnacles (Chthamalus). PLoS One 4: e5567-e5567. [ Links ]
Waters, J.M., G.A. McCulloch & J.A. Eason. 2007. Marine biogeographical structure in two highly dispersive gastropods: implications for trans-Tasman dispersal. J. Biogeogr. 34: 678-687. [ Links ]
*Correspondencia a:
Meredith K. Meyers. Odum School of Ecology, University of Georgia, Athens, Georgia, United States of America; mkmeyers@uga.edu
M. Sabrina Pankey. Department of Genetics, University of Georgia, Athens, Georgia, United States of America. Department of Ecology, Evolution, and Marine Biology, University of California Santa Barbara, Santa Barbara, California, United States of America; sabrina.pankey@lifesci.ucsb.edu.
John P. Wares. Odum School of Ecology, University of Georgia, Athens, Georgia, United States of America. Department of Genetics, University of Georgia, Athens, Georgia, United States of America; jpwares@uga.edu
1. Odum School of Ecology, University of Georgia, Athens, Georgia, United States of America; mkmeyers@uga.edu
2. Department of Genetics, University of Georgia, Athens, Georgia, United States of America; jpwares@uga.edu
3. Department of Ecology, Evolution, and Marine Biology, University of California Santa Barbara, Santa Barbara, California, United States of America; sabrina.pankey@lifesci.ucsb.edu
Received 16-I-2012. Corrected 07-VIII-2012. Accepted 09-IX-2012.