Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.61 n.2 San José Jun. 2013
An AFLP estimation of the outcrossing rate of Spondias tuberosa (Anacardiaceae), an endemic species to the Brazilian semiarid region
Una estimación AFLP de la tasa de fecundación cruzada de Spondias tuberosa (Anacardiaceae), una especie endémica de la región semiárida de Brasil
Una estimación AFLP de la tasa de fecundación cruzada de Spondias tuberosa (Anacardiaceae), una especie endémica de la región semiárida de Brasil
*Dirección para correspondencia :
Abstract
The umbu tree (Spondias tuberosa) is one of the most important endemic species to the Brazilian tropical semiarid region. The umbu tree has edible fruits with a peculiar flavor that are consumed in natura or in a semi-industrialized form, such as jams, candies and juices. The majority of endemic species to Brazilian semiarid region have not been studied or sampled to form germ- plasm collections, which increases the risk of losing genetic variability of the adapted species to xerophytic conditions. The aim of this study was to estimate outcrossing rates in S. tuberosa using a multilocus mixed model in order to guide genetic resources and breeding programs of this species. DNA samples were extracted from 92 progenies of umbu trees, which were distributed among 12 families. These trees were planted by seed in 1991 in Petrolina, PE, Brazil. The experimental design was a randomized block, with a total of 42 progenies sampled in three regions. The experimental units were composed by five plants and five replications. The out- crossing rate was estimated by the multilocus model, which is available in the MLTR software, and was based on 17 polymorphic AFLP bands obtained from AAA_CTG and AAA_CTC primer combinations. The observed heterozygotes ranged from 0.147 to 0.499, with a maximum frequency estimated for the AAA_CTC_10 amplicon. The multilocus outcrossing estimation (
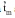 ) was 0.804±0.072, while the single-locus (
) was 0.804±0.072, while the single-locus ( 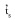 ) was 0.841±0.079, which suggests that S. tuberosa is predominantly an outcrossing species. The difference between
) was 0.841±0.079, which suggests that S. tuberosa is predominantly an outcrossing species. The difference between 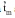 and
and 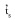 was -0.037±0.029, which indicates that biparental inbreeding was nearly absent. The mean inbreeding coefficient or fixation index (
was -0.037±0.029, which indicates that biparental inbreeding was nearly absent. The mean inbreeding coefficient or fixation index ( 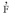 ) among maternal plants was - 0.103±0.045, and the expected
) among maternal plants was - 0.103±0.045, and the expected 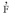 was 0.108, which indicates that there was no excess of heterozygotes in the maternal population. The outcrossing estimates obtained in the present study indicate that S. tuberosa is an open-pollinated species. Biometrical models applied to this species should therefore take into account the deviation from random outcrossing to estimate genetic parameters and the constitution of broad germplasm samples to preserve the genetic variability of the species. Outcrossing rates based on AFLP and the mixed-mating model should be applied to other studies of plant species in the Brazilian semiarid region.
was 0.108, which indicates that there was no excess of heterozygotes in the maternal population. The outcrossing estimates obtained in the present study indicate that S. tuberosa is an open-pollinated species. Biometrical models applied to this species should therefore take into account the deviation from random outcrossing to estimate genetic parameters and the constitution of broad germplasm samples to preserve the genetic variability of the species. Outcrossing rates based on AFLP and the mixed-mating model should be applied to other studies of plant species in the Brazilian semiarid region. Key words: outcrossing rate, MLTR, heterozygosis.
Resumen
El árbol de umbu (Spondias tuberosa) es una de las especies endémicas más importantes de la región semiárida del Brazil. El mismo tiene frutos comestibles con sabor distinto y puede ser consumido fresco o semiindustrializado, como mermeladas y zumos. La mayoría de las especies endémicas de la región semiárida del Brazil no fueron estudiadas o muestreadas para formar colecciones de germoplasma, aumentando el riesgo de pérdida de la variabilidad genética. El objetivo de este trabajo fue estimar las tasas de polinización cruzada en S. tuberosa basada en el modelo multi-locus mixto, con el fin de orientar los recursos genéticos y los programas de mejoramiento de esta especie. Muestras de ADN fueron extraídas de 92 progenies de árboles umbuzeiro, distribuidos en 12 familias, que se establecieron en Petrolina, PE, Brazil, 09º09’ S - 40º22’ W. El diseño experimental fue de bloques al azar con un total de 42 progenies muestreadas en tres regiones. La tasa de fecundación cruzada fue estimada por el modelo multi-locus disponible en el software MLTR, basado en 17 bandas de AFLP polimórficas obtenidas a partir de las combinaciones de cebadores AAA_CTG y AAA_CTC. Los heterocigotos observados oscilaron entre 0.147 y 0.499 con la frecuencia máxima estimada para AAA_CTC 10 amplicón. El valor estimado de cruzamiento multi-locus (
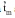 ) fue 0.804±0.072, mientras que el locus de uno-locus (
) fue 0.804±0.072, mientras que el locus de uno-locus ( 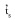 ) fue 0.841±0.079, lo que sugiere que S. tuberosa es predominantemente una especie de polinización cruzada. La diferencia entre el
) fue 0.841±0.079, lo que sugiere que S. tuberosa es predominantemente una especie de polinización cruzada. La diferencia entre el 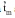 y
y 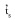 fue de -0.037± 0,029, lo que indica que la endogamia bi-parental fue casi inexistente. La media del coeficiente de fijación (
fue de -0.037± 0,029, lo que indica que la endogamia bi-parental fue casi inexistente. La media del coeficiente de fijación ( 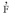 ) entre las plantas maternas fue - 0.103±0.045, mientras que la
) entre las plantas maternas fue - 0.103±0.045, mientras que la 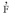 esperada fue 0.108, lo que indica que no hubo un exceso de heterocigotos en la población materna. Las estimaciones obtenidas en este trabajo indican que S. tuberosa es una especie de polinización cruzada. Los modelos biométricos aplicados a esta especie deben tener en cuenta la desviación del cruce aleatorio para estimar los parámetros genéticos y la formación de grandes muestras para preservar la variabilidad genética de esta especie. La tasa de fecundación cruzada basada en AFLP y el apareamiento mezclado debe ser aplicado a otros estudios de especies de plantas de la región semiárida del Brazil.
esperada fue 0.108, lo que indica que no hubo un exceso de heterocigotos en la población materna. Las estimaciones obtenidas en este trabajo indican que S. tuberosa es una especie de polinización cruzada. Los modelos biométricos aplicados a esta especie deben tener en cuenta la desviación del cruce aleatorio para estimar los parámetros genéticos y la formación de grandes muestras para preservar la variabilidad genética de esta especie. La tasa de fecundación cruzada basada en AFLP y el apareamiento mezclado debe ser aplicado a otros estudios de especies de plantas de la región semiárida del Brazil.Palabras clave: tasa de fecundación cruzada, MLTR, heterocigosis.
The umbu tree (Spondias tuberosa Arruda - Anacardiaceae) is a xerophytic tree species that is endemic to the tropical Brazilian semiarid region (Prado & Gibbs 1993). The high drought tolerance of this species is due to its specialized root structure (xylopodium), which plays a key role in long-term water storage. The tree has edible fruits with a peculiar flavor that are consumed in natura or semi-industrialized in different forms, such as jams, candies and juices.
This species has a greater potential for agronomic cultivation in environments with minimal rainfall (Santos 1999) or degraded soil. The umbu tree was considered an endangered species because of the harvesting of the modified roots of native plants that have reduced the native plant population. The mating system of the umbu tree has been studied using isozymes (Souza 2000), but no published studies have used co-dominant markers, such as microsatellites, or dominant markers, such as random amplification of polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP).
Understanding the mating system of a species is fundamentally important for genetic improvement and conservation programs because this information facilitates the development of strategies that optimize the sampling of genetic variability, appropriate genetic-statistical models for estimating genetic parameters, and strategies aimed at the effective conservation of species. A species can produce offspring through different types of mating, such as random outcrossing, correlated outcrossing, biparental inbreeding, self-pollination, apomixes or a combination of these reproductive strategies. Plant reproduction, together with the mechanisms of pollen and seed dispersal, determines the genetic structure of plant populations (Freitas et al. 2004).
The plant species that result from out- crossing maintain the majority of their distributed genetic variability within the population, in contrast to a predominantly self-pollinated species in which the majority of the genetic variability is distributed between populations (Hamrick & Godt 1989). Reproductive studies have revealed that a large number of plant species are allogamous or possess a mixed mating system that is predominantly allogamous (Gus- son et al. 2006).
The development and application of isozymes was a breakthrough in estimating plant population outcrossing rates and was mainly explored in perennial trees. Subsequently, DNA molecular markers, such as RAPD and AFLP, have been used to estimate the outcrossing rates of many species in order to increase the number of polymorphic markers in simple reactions (Gaiotto et al. 1997, Muluvi et al. 2004). Because of their dominance, RAPD and AFLP markers have limitations. To solve the dominance limitations, Ritland (2002) has presented a statistical estimation that is based on multilocus procedures for genes that occur in intermediate frequencies.
This study aimed to determine the dominant reproductive system of S. tuberosa using AFLP markers based on multilocus estimation in order to aid the development of strategies to sample, preserve and initiate breeding efforts in S. tuberosa.
Materials and methods
Tree samples and DNA extractions: The leaves of 92 adult S. tuberosa trees, which were planted in 1991, were sampled in the Caatinga Experimental Station of Embrapa tropical semiarid, in Petrolina, PE, Brazil, 09°09’ S - 40°22’ W. The experimental design was a randomized block, with 42 progenies sampled in three regions. The experimental units were composed by five plants and five replications. S. tuberosa progenies originated from 12 families, which numbered from five to 12 progenies/family. The collected leaves were frozen at -80ºC until DNA extraction.
Genomic DNA was extracted from the leaves using a modified Doyle & Doyle (1990) protocol: 6 000 and 10 000rpm in the first and second centrifugations, respectively; betamercaptoethanol at 2%; and incubation at 60oC for 30min. On occasion, mixing was performed by gentle tube inversion. After the addition of Tris-EDTA, the DNA solution was submitted to an RNAse treatment to remove co-isolated RNAs. The integrity and quantification of the DNA was accomplished in 0.8% agarose, followed by a DNA dilution to 40ng/μL.
Approximately 200ng of genomic DNA of each progeny was double digested for 2.5h with the endonuclease enzymes EcoRI and MseI. The digested DNA was ligated with T4 DNA ligase to a final volume of 7.25μL. Pre- amplification reactions were performed to generate a final volume of 15µL (1.5µM of each EcoRI and MseI primer, 0.2mM of each dNTP, 1x PCR buffer, 2.5mM MgCl2, 0.5 units of Taq DNA Polymerase and 2.0µL of diluted 5-fold ligated DNA solution) and were thermocycled for 20 cycles at 94oC for 30s, 56oC for 1min and then at 72oC for 1min.
After pre-amplification, the samples were diluted 20-fold in TE buffer. Selective amplifications were performed to reach a final volume of 10μL (0.2µM of EcoRI primer, 0.3µM of MseI primer, 0.2mM of dNTPs, 1x PCR buffer (100mM Tris-HCl (pH 8.3) and 500mM KCl), 2.5mM MgCl2, 0.2 units of Taq DNA polymerase and 2µL of pre-amplified DNA). The touchdown thermocycling conditions involved (a) 1 cycle at 94oC for 30s, 65oC for 30 s and 72oC for 60s; (b) 13 cycles with an initial annealing temperature of 65oC and reductions of 0.7oC for every subsequent cycle; and (c) 23 cycles at 94oC for 30s, 56oC for 30s and 72oC for 60s. A reaction of 2.0μL of formamide dye (98% formamide, 10mM EDTA, and 10mg of both bromophenol blue and xylene cyanol) was then added. The reactions were heated for 3min at 90oC and then immediately placed on ice to further allow the polyacrylamide denaturing gels to load. The gels were stained with silver nitrate according to Creste et al. (2001). All reactions were performed at the genetic laboratory of the Embrapa tropical semiarid unit.
For data analysis, the appearance of AFLP polymorphic bands was scored as a 1 for present and 0 for absent. Chi-square tests were performed for all AFLP amplicons using Mendelian inheritance ratios of 3:1, 1:1 or 1:3.
Single-locus (
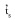 ) and multilocus
) and multilocus 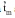 out- crossing rates, AFLP marker-allele frequencies, observed heterozygote frequencies, ovule and pollen frequencies and the coefficient of parental inbreeding (
out- crossing rates, AFLP marker-allele frequencies, observed heterozygote frequencies, ovule and pollen frequencies and the coefficient of parental inbreeding ( 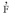 ) were estimated with the mixed-mating model (mixed outcrossing and selfing) using the software MLTR (Ritland 2002). This model specifies that both selfing and outcrossing occur in the population. One hundred bootstraps were used in the analysis. Maximum-likelihood estimates were obtained for all AFLP marker loci employed in the estimation of outcrossing rates and
) were estimated with the mixed-mating model (mixed outcrossing and selfing) using the software MLTR (Ritland 2002). This model specifies that both selfing and outcrossing occur in the population. One hundred bootstraps were used in the analysis. Maximum-likelihood estimates were obtained for all AFLP marker loci employed in the estimation of outcrossing rates and 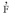 coefficients.
coefficients.Results
A total of seventeen polymorphic bands was obtained, six from the primer combination (PC) AAA_CTG and 11 from the PC AAA_CTC. These bands were easily scored in the progeny population. The bands that occurred in close positions within the gel were not scored to avoid problems in the phenotypic data.
A chi-square test at the 5% significance level indicated that of the seventeen AFLP bands, four bands (AAA_CTG_1, AAA_ CTG_2, AAA_CTC_5 and AAA_CTC_6) occurred in deviation from the Mendelian inheritance ratios of 3:1, 1:1 or 1:3.
The observed heterozygotes ranged from 0.147 to 0.499 (Table 1). The maximum frequency of heterozygotes is 0.50 (Falconer & Mackay 1996), estimated for the AFLP AAA_ CTG_3 and AAA_CTC_10 amplicons.
The multilocus outcrossing estimation (
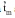 ) based on all AFLP loci was 0.804±0.072, while the single-locus estimation (
) based on all AFLP loci was 0.804±0.072, while the single-locus estimation ( 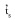 ) was 0.841±0.079. Using only the Thirteen AFLP loci that fit the Mendelian segregation (Table 1), the multilocus outcrossing estimation (
) was 0.841±0.079. Using only the Thirteen AFLP loci that fit the Mendelian segregation (Table 1), the multilocus outcrossing estimation ( 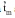 ) was 0.869±0.059, while the single-locus estimation (
) was 0.869±0.059, while the single-locus estimation ( 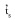 ) was 0.933±0.063.
) was 0.933±0.063.The difference between
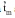 and
and 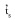 was -0.037±0.029, and the mean inbreeding coefficient or fixation index (
was -0.037±0.029, and the mean inbreeding coefficient or fixation index ( 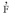 ) among the maternal plants was - 0.103±0.045.
) among the maternal plants was - 0.103±0.045.Discussion
The studied S. tuberosa population is the oldest experimental population established for genetic studies, and it contains an important gene pool for the conservation of S. tuberosa. This study matched the current efforts to conserve this important endemic species in the Brazilian semiarid region (Santos et al. 2008) as an in vivo gene bank.
The number of polymorphic bands obtained in this study was in accord with the S. tuberosa results reported by Santos et al. (2008), who reported 3 to 16 polymorphic bands from 12 AFLP PC.
The deviations from Mendelian inheritance detected in the present study were expected because all of the 92 pooled individuals were considered to be within the frequencies, and no family estimations were obtained because of the small number of progenies in some families. Pollen pool frequencies for AFLP were constrained to equal ovule frequencies by the MLTR software, therefore minimizing the violations in the mixed-mating model that were required for the correct application of the multilocus estimation.
According to Ritland (2002), when many loci are used for the estimation, it does not matter if a few loci show significant deviations because the multilocus estimate tends to be robust against violations of the assumptions. On the basis of this reasoning, the estimates on all 17 loci were considered for the present study.
The difference between
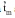 and
and 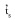 indicated that bi-parental inbreeding was nearly absent. The mean inbreeding coefficient or fixation index (
indicated that bi-parental inbreeding was nearly absent. The mean inbreeding coefficient or fixation index ( 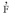 ) among maternal plants was -0.103±0.045, and the expected
) among maternal plants was -0.103±0.045, and the expected 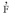 was 0.108, as calculated by
was 0.108, as calculated by 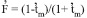 (Gaiotto et al. 1997). These results indicate that there was no excess of heterozygotes in the maternal population. An excess of heterozygotes was reported by Moraes et al. (2005), however, in Myracrodruon urundeuva, with values ranging from -0.252 to 0.511.
(Gaiotto et al. 1997). These results indicate that there was no excess of heterozygotes in the maternal population. An excess of heterozygotes was reported by Moraes et al. (2005), however, in Myracrodruon urundeuva, with values ranging from -0.252 to 0.511.The multilocus model (Ritland 2002) has a few assumptions, as revised by Sebbenn et al. (1998): (1) the pollen pool should be homogeneous in the crossing with maternal plants, (2) there should be independent segregation among the loci, (3) there should be an absence of selection and mutation, and (4) the loci should be in Hardy-Weinberg equilibrium. An assumption of intermediate allele frequencies is also expected (Gaiotto et al. 1997). Departures from these assumptions, mainly in pollen pool homogeneity, have been reported by Gaiotto et al. (1997), Sebbenn et al. (1998), Muluvi et al. (2004), Gusson et al. (2006) and Muchugi et al. (2008).
The multilocus estimation of 0.804 is close to the 0.74 value reported by Souza (2000) using three isozyme systems. in the present study, the frequencies of ovules and pollen were constrained to be equal for all AFLP markers, which suggest an absence of departures in the mixed model. These results suggest that S. tuberosa is an open-pollinated species, with a selfing rate of 0.196. S. tuberosa is an andromonoecious species and possesses hermaphrodites and male flowers in the same inflorescence. Anthesis begins at 5:00 am; hermaphrodite flowers survive for two days, and male flowers survive for one day (Nadia & Machado 2007). The multilocus model estimate is therefore in accord with the flower’s biology, as selfing occasionally occurs in this species.
As reviewed by Muchugi et al. (2008), studies on the biological characteristics of some tropical tree species have revealed high levels of outcrossing, and varying molecular marker assessments of the reproductive systems have shown high outcrossing rates. For example, Eucalyptus grandis (
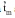 =0.84), Platypodium elegan (
=0.84), Platypodium elegan ( 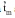 =0.92) and Shorea congestiflora (
=0.92) and Shorea congestiflora ( 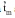 =0.87). According to the authors, outcrossing is a reasonable explanation for the high genetic variability observed in tropical trees. In an isolated study, Stacy et al. (1996) used five isozyme systems to determine that S. mombin was a completely allogamous species with 100% outcrossing.
=0.87). According to the authors, outcrossing is a reasonable explanation for the high genetic variability observed in tropical trees. In an isolated study, Stacy et al. (1996) used five isozyme systems to determine that S. mombin was a completely allogamous species with 100% outcrossing.Based on the proportion of selfing, adjustments can be made to the coefficient of additive genetic variance that is estimated from open-pollinated families when the assumption of complete half-sib relationship is not met (Gaiotto et al. 1997), as observed for S. tuberosa in the present study. The high levels of heterozygosity detected in the present study should be relevant for in situ conservation because a large number of new genetic recombinations could occur and maintain the evolutionary capacity of this species to adapt to climate changes and colonize new areas.
The outcrossing estimates obtained in the present study indicate that S. tuberosa is a predominantly open-pollinated species. Therefore, biometrical models applied to this species should take into account deviation from random outcrossing to estimate genetic parameters. As recommended by Freitas et al. (2004), while evaluating Myracrodruon urundeuva, biometrical models should also account for the constitution of broad germplasm samples to preserve the genetic variability of the species.
The high heterozygosity levels observed in the analyzed umbu tree families were also an important variable for the in situ conservation of this species because new allelic recombination can occur, thus making it possible for the appearance of new genotypes that are readily adapted to environmental changes and for the colonization of new areas.
More than 932 savanna-like plant species, including 380 endemic species, have been identified in the Brazilian semiarid Region (Brasil 2002). The majority of these species have not been studied or sampled to form a germplasm collection, which increases the risk of losing genetic variability of the adapted species to xerophytic conditions. information about the reproductive system is crucial to define a genetic resources program not only for the sampling process but also for the genetic characterization of accessions. Outcrossing rates based on AFLP and the mixed-mating model available in the software MLTR (Ritland 2002) have proven to be reliable in the present study and should be applied to other studies of plant species in the Brazilian semiarid region.
Acknowledgments
We are grateful with the Bank of Northeast of Brazil for their financial support.
References
Brasil. 2002. Ministério do Meio Ambiente: Secretaria de Biodiversidade e Florestas. Avaliação de ações prioritárias para a conservação da biodiversidade da caatinga. ibama, Brasília, Brazil. [ Links ]
Creste, S., A. Tulmann Neto & A. Figueira. 2001. Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol. Biol. Rep. 9: 299-306. [ Links ]
Doyle, J.J. & J.L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13-15. [ Links ]
Falconer, D.S. & T.F.C. Mackay. 1996. introduction to quantitative genetics. Addison Wesley Longman, Harlow, Essex, United Kingdom. [ Links ]
Freitas, M.L.M., A.M. Sebbenn, M.L.T. Moraes & E.G.M. Lemos. 2004. Mating system of a population of Myracrodruon urundeuva F.F. & M.F. Allemão using the fAFLP molecular marker. Genet. Mol. Biol. 27: 425-431. [ Links ]
Gaiotto, F.A., M. Bramucci & D. Grattapaglia. 1997. Estimation of outcrossing rate in a breeding population of Eucalyptus urophylla with dominant RAPD and AFLP markers. Theor. Appl. Genet. 95: 842-849. [ Links ]
Gusson, E., A.M. Sebbenn & P.Y. Kageyama. 2006. Sistema de reprodução em populações de Eschweilera ovata (Cambess.) Miers. Ver. Árvore. 30: 491-502. [ Links ]
Hamrick, J.L. & M.J.W. Godt. 1989. Allozyme diversity in plant species, p. 43-63. In A.H.D. Brown, M.T. Clegg, A.L. Kahler & B.S. Weir (eds.). Plant Population Genetics Breeding and Genetic Resources. Sinauer Associates, Sunderland, Massachusetts, USA. [ Links ]
Moraes, M.L.T. de, P.Y. Kageyama & A.M. Sebbenn. 2005. Diversidade e estrutura genética espacial em duas populações de Myracrodruon urundeuva Fr. All. sob diferentes condições antrópicas. Rev Árvore 29: 281-289. [ Links ]
Muchugi, A., G.M. Muluvi, A.J. Simons, F.N. Wachira & R.H. Jamnadass. 2008. Estimation of out-crossing rate in a natural breeding population of Warburgia ugandensis using AFLP marker. Afr. J. Biotechnol. 7: 139-146. [ Links ]
Muluvi, G.M., J.I. Sprent, D. Odee & W. Powell. 2004. Estimates of outcrossing rates in Moringa oleifera using Amplified fragment length polymorphism (AFLP). Afr. J. Biotechnol. 3: 146-151. [ Links ]
Nadia, T. de L. & I.C. Machado. 2007. Polinização de Spondias tuberosa Arruda (Anacardiaceae) e análise da partilha de polinizadores com Ziziphus joazeiro Mart. (Rhamnaceae), espécies frutíferas e endêmicas da caatinga. Ver. Bras. Bot. 30: 89-100. [ Links ]
Prado, D.E. & P.E. Gibbs. 1993. Patterns of species distribution in the dry seasonal forests of South America. Ann. Mo Bot Gard 80: 902-927. [ Links ]
Ritland, K. 2002. Extensions of models for the estimation of mating systems using n independent loci. Heredity 88: 221-228. [ Links ]
Santos, C.A.F. 1999. in situ evaluation of fruit yield and estimation of repeatability coefficient for major fruit traits of umbu tree (Spondias tuberosa (Anacardiaceae) in the semi-arid region of Brazil. Genet. Resour Crop. Ev. 46: 455-460. [ Links ]
Santos, C.A.F., M.A. Rodrigues & M.i. Zucchi. 2008. Variabilidade genética do umbuzeiro no Semi-Árido brasileiro por meio de marcadores AFLP. Pesqui. Agropecu. Bras. 43: 1037-1043. [ Links ]
Sebbenn, A.M., P.Y. Kageyama & R. Vencovsky. 1998. Variabilidade genética, sistema reprodutivo e estrutura genética espacial em Genipa americana L. através de marcadores isoenzimáticos. Sci. For. 53: 15-30. [ Links ]
Souza, J.C. de. 2000. Variabilidade genética e sistema de cruzamento em populações naturais de umbuzeiro (Spondias tuberosa Arr. Cam.). Ph.D. Thesis, Univer- sidade Federal de Viçosa, Brazil. [ Links ]
Stacy, E.A., J.L. Hamrick, J.D. Nason, S.P. Hubbeell, R.B. Foster & R. Condit. 1996. Pollen dispersal in low- density populations of three neotropical tree species. Am. Nat. 148: 275-298. [ Links ]
*Correspondencia:
Carlos Antonio Fernandes Santos:Embrapa Semiárido, Caixa Postal 23. 56300-970. Petrolina, PE. Brazil; casantos@cpatsa.embrapa.br
Renata Natália Cândido de Souza Gama: Universidade Estadual de Feira de Santana. Avenida Transnordestina, SN, Novo Horizonte, 44.036-900, Feira de Santana-BA, Brazil; renata.natalia@hotmail.com
1. Embrapa Semiárido, Caixa Postal 23. 56300-970. Petrolina, PE. Brazil; casantos@cpatsa.embrapa.br
2. Universidade Estadual de Feira de Santana. Avenida Transnordestina, SN, Novo Horizonte, 44.036-900, Feira de Santana-BA, Brazil; renata.natalia@hotmail.com
eceived 10-I-2012. Corrected 10-IX-2012. Accepted 09-X-2012.














