Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.61 n.1 San José Mar. 2013
Cultivable bacterial diversity along the altitudinal zonation and vegetation range of tropical Eastern Himalaya
*Dirección para correspondencia:
Abstract
The Northeastern part of India sprawls over an area of 262 379km2 in the Eastern Himalayan range. This constitutes a biodiversity hotspot with high levels of biodiversity and endemism; unfortunately, is also a poorly known area, especially on its microbial diversity. In this study, we assessed cultivable soil bacterial diversity and distribution from lowlands to highlands (34 to 3 990m.a.s.l.). Soil physico-chemical parameters and forest types across the different altitudes were characterized and correlated with bacterial distribution and diversity. Microbes from the soil samples were grown in Nutrient, Muller Hinton and Luria-Bertani agar plates and were initially characterized using biochemical methods. Parameters like dehydrogenase and urease activities, temperature, moisture content, pH, carbon content, bulk density of the sampled soil were measured for each site. Representative isolates were also subjected to 16S rDNA sequence analysis. A total of 155 cultivable bacterial isolates were characterized which were analyzed for richness, evenness and diversity indices. The tropical and sub-tropical forests supported higher bacterial diversity compared to temperate pine, temperate conifer, and sub-alpine rhododendron forests. The 16S rRNA phylogenetic analysis revealed that Firmicutes was the most common group followed by Proteobacteria and Bacteroidetes. Species belonging to the genera Bacillus and Pseudomonas were the most abundant. Bacterial CFU showed positive but insignificant correlation with soil parameters like pH (r=0.208), soil temperature (r=0.303), ambient temperature (r=0.443), soil carbon content (r=0.525), soil bulk density (r=0.268), soil urease (r=0.549) and soil dehydrogenase (r=0.492). Altitude (r=0.561) and soil moisture content (r=-0.051) showed negative correlation. Altitudinal gradient along with the vegetation and soil physico-chemical parameters were found to influence bacterial diversity and distribution. This study points out that this is a biome with a vast reservoir of bacteria which decrease with increasing altitudes, and highlights the microbiological importance of the poorly studied Eastern Himalayan range, justifying efforts to explore the prevalence of novel species in the biome.
Key words: cultivable bacteria, Eastern Himalyan range, tropical region, altitude, 16S rRNA, diversity indices.
Resumen
La parte noreste de la India se extiende sobre una superficie de 262 379km2 en la cordillera oriental del Himalaya. Es un punto de acceso con altos niveles de biodiversidad y endemismo; desafortunadamente, también es una zona poco conocida, sobre todo su diversidad microbiana. En este estudio se evaluó la diversidad de bacterias cultivables del suelo, su diversidad y distribución de las tierras bajas a las altas (34 a 3 990m.s.n.m). Se caracterizaron los parámetros físico-químicos del suelo y tipos de bosques a lo largo del gradiente altitudinal y se correlacionaron con la distribución y diversidad bacteriana. Los microbios del suelo se cultivaron en placas de agar enriquecido Muller Hinton y Luria-Bertani, e inicialmente se caracterizaron mediante métodos bioquímicos. Parámetros tales como actividad de la deshidrogenasa y ureasa, temperatura, contenido de humedad y de carbono, pH y densidad aparente del suelo se midieron en cada sitio. Aislamientos representativos también se sometieron al análisis secuencial de 16S rADN. Un total de 155 aislamientos bacterianos cultivables se caracterizaron para estimar los índices de riqueza, equidad y diversidad. Los bosques tropicales y subtropicales albergan una mayor diversidad bacteriana en comparación con los bosques templados de pino y coníferas, y los bosques subalpinos de rododendro. El análisis filogenético de 16S rARN reveló que Firmicutes fue el grupo más común, seguido de Proteobacteria y Bacteroidetes. Especies pertenecientes a los géneros Bacillus y Pseudomonas fueron las más abundantes. Las UFC bacterianas mostraron una positiva pero insignificante correlación con los parámetros del suelo, tales como pH (r=0.208), temperatura (r=0.303), temperatura ambiente (r=0.443), contenido de carbón (r=0.525), densidad aparente (r=0.268), ureasa (r=0.549) y deshidrogenasa (r=0.492). La altitud (r=-0.561) y el contenido de humedad del suelo (r=-0.051) mostraron una correlación negativa. Se encontró que el gradiente altitudinal, junto con la vegetación y los parámetros físico-químicos influyeron en la diversidad bacteriana y la distribución. Este estudio señala que este es un bioma con un vasto reservorio de bacterias que disminuyen con la altitud y pone en relieve la importancia microbiológica de la pobremente estudiada zona del este del Himalaya, lo que justifica los esfuerzos para explorar la prevalencia de nuevas especies en el bioma.
Palabras clave: bacterias cultivables, zona del este del Himalaya, región tropical, altitud, 16S rARN, índices de diversidad.
The Eastern Himalayan range, one of the biodiversity hotspots (Myers et al. 2000), is facing imminent threat due to anthropogenic interventions through tourism, agriculture, industrial activities and housing. This hotspot known to harbor plants and animals with high levels of biodiversity and endemism, is poorly understood as far as microbial diversity is concerned. Despite its high potential for biodiversity conservation, Eastern Himalayan range has attracted little attention from scientists and conservationists. As strategies for conserving species and communities focuses as much on species richness and endemism (Lovett et al. 2000, Myers et al. 2000, Kier & Barthlott 2001), information about the microbiologically unexplored Eastern Himalayan range would greatly increase the potential of finding novel species (Kapur & Jain 2004).
Soil microbiota plays important roles in soil aggregation and cycling of nutrients through cellulose and lignin breakdown, and nitrogen fixation (Donnelly et al. 1990, Hu & van Bruggen 1997, Atlas & Bartha 2002). The microbial diversity of soil is huge (Alexander 1977, Rondon et al. 1999, Ward 2002) and hence, a representative estimate of microbial diversity is a prerequisite for understanding the functional activity of microorganisms in ecosystems (Garland & Mills 1994, Zak et al. 1994). The role of soil enzymes is well established by way of relationships among soil enzymes, the environmental factors and biological transformations important to soil fertility (Moore & Russell 1972, Paul & Mclaren 1975, Tabatabai 1977, Bremner & Mulvaney 1978, Brzezinska et al. 1998, Baum et al. 2003, Makoi & Ndakidemi 2008, Gao et al. 2010, Sumathi et al. 2012). Our knowledge of soil microbial diversity is limited in part by our inability to culture them and other by the lack of survey in certain areas of the Earth. However, surveying of 16S rRNA genes targeted by primers in soils, as well as other techniques like metagenomics, has permitted a more direct census of studying soil microbial diversity (Kirk et al. 2004, Palmer et al. 2006).
The immense variety of the climatic, edaphic and altitudinal variations have resulted in a great range of diverse and extreme ecological habitats in the Eastern Himalayan range. Lying between 22º11’190’’ - 28º23’261’’ N and 89º86’821’’ - 97º42’683’’ E, and sprawling over 262 379km2, Northeastern region of India represents the transition zone between the Indian, Indo-Malayan and Indo-Chinese biogeographic regions and is a meeting place of the Himalayan mountains with peninsular India. The region occupies 7.7% of India’s total geographical area and supports a rich biodiversity spanning from tropical evergreen forests, temperate conifer, to sub-alpine rhododendron forests. The faunal and floral diversity of this region (Chatterjee et al. 2006) including the mountain forests of Asia (Culmsee et al. 2011) have been well studied. However, information on microbial diversity from this range is relatively sparse and no comprehensive study has so far been made to explore and document the bacterial diversity of the region as a whole.
Most studies of bacterial diversity in soils have focused on inland desert or coastal locations (vishniac 1993). Investigations of bacteria in extreme environments such as the Antarctic region soils have focused on abundance and diversity and the influence of climatic conditions. These bacteria were assigned to known genera including Arthrobacter, Micrococcus, Bacillus and Pseudomonas (vishniac 1993). This study is the first attempt to assess the bacterial diversity along the high altitudes of the Eastern Himalayan range under different forests types and soil physico-chemical conditions. This aimed at generating a baseline data on the cultivable bacterial diversity from this unique place of the world.
Materials and Methods
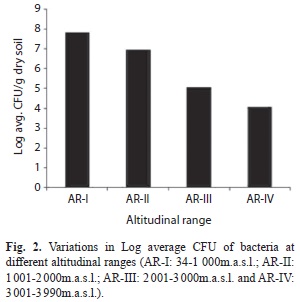
Research area: Northeast India in the Eastern Himalayan range is lying between 22º11’190’’ - 28º23’261’’N and 89º86’821’’ - 97º42’683’’E, and sprawling over 262 379km2; this represents the transition zone between the Indian, Indo-Malayan and Indo-Chinese biogeographic regions, and a meeting place of the Himalayan mountains and peninsular India, characterized by a great range of ecological habitats under different forests types, that were selected for the present study. Soils from 40 different locations covering different altitudes and having different vegetation types (Champion & Seth 1968) were sampled and analyzed during 2008 to 2011. Based on the altitude having specific vegetation and forest types, the sites were grouped into four altitudinal ranges viz., AR-I (34m.a.s.l to 1 000m.a.s.l.), AR-II (1 001m.a.s.l. to 2 000m.a.s.l.), ARIII (2 001m.a.s.l. to 3 000m.a.s.l.) and AR-Iv (3 001m.a.s.l. to 3 990m.a.s.l.).
Sample collection and sampling site parameters: Soil samples were collected using a sterilized soil corer (inner diameter 5.5cm) from a depth of 10-30cm. In order to document maximum bacterial population and diversity, five soil samples were selected from each location and the samples were pooled before analysis. The soil samples were kept in sterile containers and stored at 4ºC until processing within 24h (Joshi et al. 1991). The geographical location of each sample collection site was recorded using digital GPS (Garmin 7.6).
Soil parameters: The soil temperature was measured on site by using a soil thermometer. Soil pH was measured using a soil-water mixture 1:5 (w/v) with a pH meter. Soil moisture content was determined gravimetrically by oven drying 10g of fresh sieved soil for 24h at 105ºC (Anderson & Ingram 1993). Furthermore, bulk density was determined by Blake & Hartge (1986) method using soil corer, while soil organic carbon was determined by using Walkley & Black’s rapid titration method as described by Anderson & Ingram (1993).
Soil Dehydrogenase activity: The dehydrogenase is used as a general criterion to determine soil microbial activity and is considered a good measure of soil microbial oxidation activity; soil bacterial dehydrogenase activity was estimated following the method as described by Kumar (2011). After incubating at 37ºC for 24h, the formazan formed was extracted with 10mL ethanol and estimated spectrophotometrically at 485nm. The formazan concentration was calculated from its standard curve. The dehydrogenase activity is expressed as gram of formazan formed per gram of dry weight of soil.
Soil Urease activity: Soil enzymes activities including urease activities are sensors of soil degradation since they integrate information about microbial status and soil physico-chemical conditions (Wick et al. 1998, Aon & Colaneri 2011, Baum et al. 2003); the urease test has also been used to detect bacteria from soil (Zaved et al. 2008). Urease activity in soils was assayed by the buffer method (Tabatabai 1994) which involves the determination of the NH4+ released when a soil sample is incubated with THAM (Tris hydroxymethyl aminomethane) buffer at the optimal pH (pH 9.0) with or without toluene, and urea (0.2M) at 37ºC for 2h.
Isolation, enumeration and preservation of bacteria: The total colony forming units (CFU) of cultivable soil bacteria were determined using a soil dilution plate-count technique. A total of 10g of soil samples were suspended in 100mL sterile 0.85% NaCl solution and mixed by shaking in a shaker incubator at 120rpm for 4-5min to establish a dilution series; these were used as inoculum for plate count of colony forming units (CFU). Aliquots of 100μL from different dilutions were transferred and spread onto Nutrient agar, Muller Hinton agar and Luria-Bertani agar plates in triplicates separately. Three different culture media were used in order to record maximum diversity of cultivable bacteria; however, nutrient agar was considered as the standard media based on the isolation efficiency. The agar plates were incubated at 35-37ºC for 24-48h and colonies formed on the plates were counted for enumeration. Well-isolated colonies with different morphologies were randomly chosen from each plate and streaked on fresh plates. Isolates were checked for purity by re-streaking and then inoculated into nutrient agar slants and stored at 4ºC. Simultaneously, the pure cultures of isolates were preserved in 20% glycerol water at -20ºC (Kumar et al. 2011).
Growth and preliminary characterization: Colony morphology was determined after two to seven days growth on agar plates incubated at 35-37ºC. Each isolate was subjected to Gram staining and was examined for cellular morphology and arrangement. various tests for biochemical characterization were performed following standard protocols given by Holt et al. (1994).
Molecular characterization: A total of 155 selected isolates were subjected to 16S rDNA sequence analysis following the methods as described by Kumar et al. (2011). Briefly, Genomic DNA was extracted using Genomic DNA isolation kit (HiPurA Bacterial and Yeast DNA Purification Spin Kit, HiMedia, India). The 16S rRNA gene sequences were amplified by Gene AMP 9700 (Applied Biosystems, USA) PCR using two universal bacterial 16S rRNA gene primers (i.e., 27F 5’-AGAGTTTGATCCTGGCTCAG-3’ and 1492R 5’-TACGGYTACCTTGTTACGACTT-3’). PCR mixtures (25μL) contained approximately 30ng of DNA, 2μM each forward primer 27F and reverse primer 1492R, 1.5mM of MgCl2 (Taq Buffer), deoxynucleoside triphosphates (250μM each of dATP, dCTP, dGTP and dTTP) and 0.6U of Taq polymerase. PCR cycle used for the reaction comprised an initial denaturation for 5min at 94ºC, and this was followed by 30 cycles each of denaturation at 94ºC for 1min, annealing at 55ºC for 1min, and extension at 72ºC for 2min, and the final extension for 5min at 72ºC. Approximately, 1 500 nucleotides were amplified. For negative controls sterile distilled water was used in place of DNA template. Amplified products were purified using QIAquick Gel Extraction Spin Kit (QIAGEN, Germany).
The purified PCR products were bi-directionally sequenced by Genetic Analyzer ABI 3130xL (Applied Biosystems, California, USA) with Big Dye (3.1) Terminator protocol using the forward, reverse and internal primers corresponding to Escherichia coli positions 357F, 926F, 685R and 1100R. Sequencing reaction was performed with 20μL reaction mixture containing approximately 50ng of template DNA and 1pmol of sequencing primers. Post reaction cleanup and resuspension were performed for removal of unincorporated dye terminators from the sequencing reaction using 125mM EDTA, 3M sodium acetate and 70% ethanol. Basic Local Alignment Search Tool (BLAST) was used to determine the phylogenetic neighbors from the nucleotide database of National Centre for Biotechnology Information (NCBI) (Altschul et al. 1997) and EzTaxon (the database of type strains with validly published prokaryotic names available online at http://www.eztaxon.org/) (Chun et al. 2007).
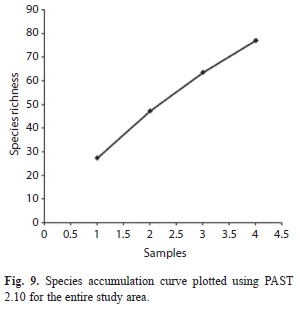
The correlation between the different environmental factors, soil physico-chemical parameters and bacterial counts was determined by calculating Pearson product moment correlation coefficients (Zar 1984). The correlations were considered significant if p<0.05. Karl Pearson Correlation Coefficient (Parametric test) was done using Minitab 11.5 Statistical Software to study the influence of soil physico-chemical and environmental parameters on soil bacterial distribution/diversity. The diversity of the bacterial community was calculated using the Shannon-Wiener’s (H’) method (Jost 2006). However, the entropy value (H’) of the Shannon index is not itself a measure of diversity. Conversion of this value to effective number of species, or true Diversity (D), is the key to a unified and intuitive interpretation of diversity (Jost 2006). The complement of Simpson’s index, (d’) (Krebs 1978) was also estimated. Bacterial alpha diversity was estimated with Fisher’s α (Magurran 2004). Evenness (J) of species was evaluated using the formula as given by Pielou (1966). The number of species divided by the square root of the number of individuals results in Species richness, S. Species accumulation curve (Colwell et al. 2004), show the rate at which new species are found within a community and can be extrapolated to provide an estimate of species richness. It was plotted using PAST 2.10 software (Hammer et al. 2001).
Results
Research area and soil physico-chemical properties: Tropical and sub-tropical forests were prevalent at lower altitudes whereas temperate pine to temperate conifer and sub-alpine rhododendron forests were abundant at higher altitudes (Table 1). The prevalent vegetations, ambient temperature, soil pH, soil temperature, soil moisture content, soil carbon content and bulk density were found to vary along the altitudes (Table 1). Soil physico-chemical factors, ambient temperature and/or substrate availability were found to influence soil bacterial growth and population density at various level of significance (p<0.05) (Fig. 1). Soil carbon was found to have significant positive correlation with soil temperature (r=0.785, p<0.05) (Fig. 1).
Enzymes activity: The enzymes activities showed negative correlation with altitude, as highest activities for both urease and dehydrogenase enzymes were recorded at lower altitudes and vice versa (Table 1). Urease activity ranged from 0.09 to 0.28 (SD=±0.0508) (mg) NH4+/g of dry soil, whereas that of dehydrogenase varied from 0.07 to 0.16 (SD=±0.04788) (mg) TPF/g of dry soil. A significant positive correlation was observed between soil dehydrogenase and urease activity. Soil carbon was found to have significant positive correlation with urease and dehydrogenase activity (Fig. 1).
Cultivable bacteria: The bacterial colonies observed on agar plates ranged from 1.8x103 to 3.2x108 (SD=±6.3x107) cfu/g of dry soil. There was significant variation in the soil physico-chemical parameters and bacterial CFU counts along the altitudinal gradient (Table 1). The CFU counts of bacteria decreased with increasing altitude, as highest CFU counts were recorded at lower altitudes and lowest CFU counts were recorded at higher altitudes (Table 1, Fig. 2). Soil bacterial distribution showed positive but insignificant correlation with soil pH, soil temperature, ambient temperature, soil carbon content, soil bulk density, urease and dehydrogenase activities whereas altitude and soil moisture content showed negative correlation (Fig. 1).
Phylogenetic diversity of bacterial isolates: A total of 155 bacterial isolates (Table 2) were obtained using cultivation based method from the entire altitude (34-3 990m.a.s.l) under the study area. Characterized isolates belonged to Proteobacteria, Firmicutes and Bacteroidetes, which comprised of Alpha-, Betaand Gamma-Proteobacteria, Bacilli and Flavobacteria (Table 3), distributed to 27 genera and 77 species. From among the 155 isolates; 103 belonged to Firmicutes, 47 to Proteobacteria (two to α-Proteobacteria, 34 to γ-Proteobacteria and 11 to β-Proteobacteria) and five to Bacteroidetes. The phylogentic trees for the respective groups (Betaand Gamma-Proteobacteria, Firmicutes and Flavobacteria) are constructed using rooted Neighbor-joining tree based on 16S rRNA (more than 1,400 bases) gene sequences of the isolates and the related species obtained from the database of type strains with validly published prokaryotic names at the EzTaxon server. The scale bars given in the phylogenetic trees correspond to the expected number of changes per nucleotide position (Figs. 3-6C).
The predominant phyla characterized in the entire stretch of altitudes were Firmicutes followed by Proteobacteria and Bacteroidetes (Fig. 7). These three phyla were found to occur in all the four altitudinal ranges with varying density. Under Proteobacteria, β-Proteobacteria and γ-Proteobacteria were isolated from all the four altitudinal ranges whereas α-Proteobacteria was isolated only from AR-III and AR-Iv. Bacillus and Lysinibacillus were the dominant genus under the class Bacilli. Similarly, Serratia and Pseudomonas belonging to class γ-Proteobacteria were frequently characterized from all the altitudinal ranges. Under β-Proteobacteria, Burkholderia and Chromobacteria were dominant. Sphingobium and Ponticoccus were the isolated genera under α-Proteobacteria. Chryseobacterium was the only genus identified under Flavobacteria (Table 3). The genus Bacillus was the most dominant among all the isolated genera followed by Pseudomonas and Lysinibacillus. Similarly, diversity index of genus Bacillus (0.353) was found to be the highest followed by Pseudomonas (0.242) and Lysinibacillus (0.176) (Fig. 8).
Diversity indices and data analyses: Shannon-Wiener’s index (H’), Diversity (D), Complement of Simpson’s index (d’), Fisher’s alpha index, richness and evenness index for the studied range was found to be 3.92, 50.40, 0.966, 60.77, 6.18 and 0.903, respectively (Table 4). The Shannon-Wiener’s index (H’) for the entire range varied from 2.99 to 3.35. The calculated values for Shannon-Wiener’s, Diversity, complement of Simpson’s indices and species richness decreased with increasing altitude. However, Fisher’s alpha did not follow the same trend of diversity (Table 4). The highest value for most of the diversity indices as well as species richness was recorded for the lowest altitudinal range AR-I (H’=3.35, D=28.50, d’=0.955, S=4.81). Genus wise, the individual diversity index varied from 0.033 to 0.353 with Bacillus (0.353) having the highest diversity index, followed by Pseudomonas 0.242) and Lysinibacillus (0.177) (Fig. 8). The species accumulation curve displayed an increasing trend that did not level off (Fig. 9).
Accession numbers of nucleotide sequences: The 16S rDNA nucleotide partial sequences were submitted to GenBank and accessions were obtained (Table 2).
Discussion
Ecological factors and altitudinal gradient are known to influence distribution of soil micro-organisms (Lomolino 2001). The cultivable bacterial diversity across the altitude showed variation in the region revealing spatial trend and correlation with soil parameters and forest types. The variability in population densities of cultivable soil bacteria can be attributed to soil properties, physico-chemical conditions and vegetations which are among the most important factors that influence soil microbial growth, population density and diversity (Atlas 1984, Dimitriu & Grayston 2010). Alpine and sub-alpine regions situated at higher altitude with sparse vegetation represent different soil physico-chemical factors when compared to tropical and sub-tropical evergreen or deciduous forests at lower altitudes. Tropical and sub-tropical forest prevalent at lower altitudes supported higher bacterial diversity compared to temperate pine, temperate conifer and sub-alpine rhododendron forests at higher altitudes as measured by Shannon-Wiener’s (H’), Complement of Simpson’s (d’) indices and Diversity (D). This variation in bacterial diversity may be attributed to the types of forests as vegetations are known to affect soil microbial diversity and community structures (Kowalchuk et al. 2002, Grayston & Prescott 2005, Han et al. 2007, Thomson et al. 2010). The value of the Shannon-Wiener’s Index (H’) usually lies between 1.5 and 3.5 for ecological data and rarely exceeds 4.0 (May 1975). In this study, we found that the entire study area has high value of H’ (H’>3.5; Table 4) and of Fisher’s α suggesting prevalence of diversity in the region.
Richness and evenness of bacterial communities reflect selective pressure that shape diversity within communities. Measuring these parameters is most useful when assessing the relationships of soil physico-chemical and environmental parameters on bacterial diversity in soil (Kapur & Jain 2004). However, in the present study correlation was found to be insignificant. Similar findings have been reported by Bryant et al. (2008) and Fierer et al. (2011). The insignificant decrease in diversity with altitude is attributed to higher bacterial diversity at sub-tropical and temperate soil. One of the possible reasons for higher bacterial diversity at intermediate elevation could be due to the fact that the trees of sub-tropical and temperate regions including Cedrus, Pinus and Taxus in Himalayan range are known to exert slightly stimulatory effect on the microorganisms and therefore support relatively higher microbial population (Pandey et al. 2006). Similar effects of vegetations on soil microbial diversity and community structures has also been reported by several other workers (Kowalchuk et al. 2002, Grayston & Prescott 2005, Han et al. 2007, Thomson et al. 2010). The present study corroborates with the findings of Collins & Cavigelli (2003) and Fierer et al (2011) who had reported decrease in soil pH with increase in altitude. Insignificant correlation between soil temperature and bacterial diversity could be due to the variation in sampling time and season.
Members of the phyla Proteobacteria and Firmicutes are the most abundant soil bacteria, as revealed by analysis of 16S rRNA gene (Janssen 2006, Bruce et al. 2010, Lin et al. 2010). In the present study, γ-Proteobacteria was found to be more common as compared to β-Proteobacteria, which could be due to the soil physico-chemical factors as pH is found to influence the distribution of Proteobacteria (Lin et al. 2010). The abundance of Burkholderia species among β-Proteobacteria is due to its nutritional versatility as saprophyte and also being a common resident of rhizosphere soil (Estrada-De los Santos et al. 2001, Coenye & vandamme 2003). Within the Firmicutes, 75.19% of the isolates belonged to Bacillus. The high number and high diversity index value of Bacillus as compared to the other isolated genera in the study suggests that aerobic or facultative anaerobic, spore-forming bacteria are abundant in the forest soils of this Himalayan range which supports the patterns observed in temperate and tropical soils (Teixeira et al. 2010). Abundance of Serratia and Bacillus species is attributed to their capability of adapting to a wide range of environmental conditions. Both are distributed widely in nature and are commonly found in soil as saprophytic organisms (vilain et al. 2006). Plants benefit from the presence of B. cereus since it is capable of inhibiting plant diseases and also enhances plant growth (Jensen et al. 2003). Serratia marcescens, an enterobacteria, on the other hand has a unique ability to produce extracellular enzymes. Several such enzymes have been shown to have the ability to degrade chitin, a substance which mainly comprises fungal cell walls (Hejazi & Falkiner 1997). Lysinibacillus xylanilyticus is a xylan-degrading soil bacteria widely found in forest soil (Lee et al. 2010). B. aryabhattai is recorded in large number in the present study and its abundance in soil indicate good soil health as it performs wide range of enzymatic activities and play active role in soil nutrient decomposition and mineralization (Shivaji et al. 2009, Yadav et al. 2011). Bacillus from high altitudes of Eastern Himalayan range has been reported to produce thermostable enzyme (Devi et al. 2010). The cultivation approach showed that certain isolates belonging to Bacillus, Brevibacillus, Lysinibacillus and Enterobacter spp. may represent novel species as they have relatively low
16S rRNA sequence similarity≤97% (Table 2) to the known species.
Determination of dehydrogenase and urease activity in general is a criterion used to determine soil microbial activity (Burns 1978, Makoi & Ndakidemi 2008). Dehydrogenase activity is an indicator of potential non-specific intracellular enzyme activity of the total microbial biomass and may be considered a good measure of microbial oxidative activities in soils (Subhani et al. 2001).
The present study is the first broad survey on the bacterial community structure in the Eastern Himalayan soils under different vegetations and soil physico-chemical conditions. This study clearly points out that this biome represents a vast reservoir for bacterial discovery. Moreover, the species accumulation curve showed that the rate of species increase with sampling effort had not yet reached an asymptote, indicating that the diversity of the area had not yet been fully captured. The bacterial diversity data may become a baseline to compare how anthropogenic and climatic interventions in the coming years change the composition and diversity of bacteria across the altitudes of this anthropogenically exposed range. Further functional analyses are required for understanding the functional diversity (Hollister et al. 2010) and its possible exploration for bioprospection potentials and conservation value of the microbiota. Unless the area is protected, human generated disturbances such as tourism, agriculture, industrial activities, transient farming and housing may rapidly reduce the forest cover and with it, the associated microbial biodiversity value of the region in the near future, by exerting severe impacts on its rich but poorly known microflora. Future efforts focused on deep sequencing of soil microbial diversity and novel characterization methodologies to recover and describe taxonomically diverse novel species of this range are required to confirm these first conclusions, and thereby add data needed to support decisions on conservation and sustainable utilization of this biodiversity hotspot.
Acknowledgment
NAL, KK and SRJ acknowledge the financial support received from Department of Information Technology (MC & IT), Government of India to carry out the present study.
References
Alexander, M. 1977. Introduction to soil microbiology. John Wiley & Sons, New York, New York, USA. [ Links ]
Altschul, S.F., T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller & D.J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [ Links ]
Anderson, J.M. & J.S.I. Ingram. 1993. Tropical soil biology and fertility: A Handbook of Methods. CAB International, Wallingford, Washington, USA. [ Links ]
Aon, M.A. & A.C. Colaneri. 2001. II. Temporal and spatial evolution of enzymatic activities and physico-chemical properties in an agricultural soil. Appl. Soil Ecol. 18: 255-270. [ Links ]
Atlas, R.M. 1984. Diversity of microbial communities, p. 1-47. In K.C. Marshall (ed.). Advances in Microbial Ecology. Plenum, New York, New York, USA. [ Links ]
Atlas, R. & R. Bartha. 2002. Ecología microbiana y microbiología ambiental. Pearson Educación, Madrid. [ Links ]
Baum, C., P. Leinweber & A. Schlichting. 2003. Effects of chemical conditions in re-wetted peats on temporal variation in microbial biomass and acid phosphatase activity within the growing season. Appl. Soil Ecol. 22: 167-175. [ Links ]
Blake, G.R. & K.H. Hartge. 1986. Bulk Density, p. 363-375. In A. Klute (ed.). Methods of soil analysis, part I. Physical and mineralogical methods: Agronomy Monograph. American Society of Agronomy, Madison, USA. [ Links ]
Bremner, J.M. & R.L. Mulvaney. 1978. Urease activity in soils, p. 149-196. In R.G. Burns (ed.). Soil Enzymes. Academic, London, United Kingdom. [ Links ]
Bruce, T., I.B. Martinez, O.M. Neto, A.C.P. vicente, R.H. Kruger & F.L. Thompson. 2010. Bacterial community diversity in the Brazilian Atlantic forest soils. Microb. Ecol. 60: 840-849. [ Links ]
Bryant, J.A., C. Lamanna, H. Morlon, A.J. Kerkhoff, B.J. Enquist & J.L. Green. 2008. Microbes on mountain slides: Contrasting elevation pattern of bacterial and plant diversity. PNAS 105: 11505-11511. [ Links ]
Brzezinska, M., Z. Stepniewska & W. Stepniewski. 1998. Soil oxygen status and dehydrogenase activity. Soil Biol. Biochem. 30: 1783-1790. [ Links ]
Burns, R.G. 1978. Enzvme activitv in soil: Some theoretical and practical considerations, p. 295-340. In R.G. Burns (ed.). Soil Enzymes. Academic, London, United Kingdom. [ Links ]
Champion, H.G. & S.K. Seth. 1968. A revised survey of the forest types of India. Govt. of India, Delhi, India. [ Links ]
Chatterjee, S., A. Saikia, P. Dutta, D. Ghosh & S. Werah. 2006. Biodiversity significance of North east India. WWF India, Delhi, India. [ Links ]
Chun, J., J.H. Lee, Y. Jung, M. Kim, S. Kim, B.K. Kim & Y.W. Lim. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57: 2259-2261. [ Links ]
Coenye, T. & P. vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5: 719-729. [ Links ]
Collins, H.P. & M.A. Cavigelli. 2003. Soil microbial community characteristics along an elevation gradient in the Laguna Mountains of Southern California. Soil Biol. Biochem. 35: 1027-1037. [ Links ]
Colwell, R.K., C.x. Mao & J. Chang. 2004. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology 85: 2717-2727. [ Links ]
Culmsee, H., R. Pitopang, H. Mangopo & S. Sabir. 2011. Tree diversity and phytogeographical patterns of tropical high mountain rain forests in Central Sulawesi, Indonesia. Biodivers. Conserv. 20: 1103-1123. [ Links ]
Devi, S.L., P. Khaund & S.R. Joshi. 2010. Thermostable α-amylase from natural variants of Bacillus spp. prevalent in eastern Himalayan range. Afr. J. Microb. Res. 4: 2534-2542. [ Links ]
Dimitriu, P.A. & S.J. Grayston. 2010. Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb. Ecol. 59: 563-573. [ Links ]
Donnelly, P.K., J.A. Entry, D.L. Crawford & Jr. K. Cromack. 1990. Cellulose and lignin degradation in forest soils: response to moisture, temperature, and acidity. Microb. Ecol. 20: 289-295. [ Links ]
Estrada-De los Santos, P., R. Bustillos-Cristales & J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67: 2790-2798. [ Links ]
Fierer, N., C.M. McCain, P. Meir, M. Zimmermann, J.M. Rapp, M.R. Silman & R. Knight. 2011. Microbes do not follow the elevation diversity pattern of plant and animals. Ecology 92: 797-804. [ Links ]
Gao, Y., P. Zhou, L. Mao, Y. Zhi, C. Zhang & W. Shi. 2010. Effects of plant species coexistence on soil enzyme activities and soil microbial community structure under Cd and Pb combined pollution. J. Environ. Sc. (China). 22: 1040-1048. [ Links ]
Garland, J.L. & A.L. Mills. 1994. A community-level physiological approach for studying microbial communities, p. 77-83. In K. Ritz, J. Dighton & K.E. Giller (eds.). Beyond the biomass. John Wiley & Sons, Chichester, United Kingdom. [ Links ]
Grayston, S.J. & C.E. Prescott. 2005. Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol. Biochem. 37: 1157-1167. [ Links ]
Han, X.M., R.Q. Wang, J. Liu, M.C. Wang, J. Zhou & W.H. Guo. 2007. Effects of vegetation type on soil microbial community structure and catabolic diversity assessed by polyphasic methods in North China. J. Environ. Sci. 19: 1228-1234. [ Links ]
Hammer, Ø., D.A.T. Harper & P.D. Ryan. 2001. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4: 1-9. [ Links ]
Hejazi, A. & F.R. Falkiner. 1997. Serratia marcescens. J. Med. Microbiol. 46: 903-912. [ Links ]
Hollister, E.B., C.W. Schadt, A.v. Palumbo, R.J. Ansley & T.W. Boutton. 2010. Structural and functional diversity of soil bacterial and fungal communities following woody plant encroachment in the southern Great Plains. Soil Biol. Biochem. 42: 1816-1824. [ Links ]
Holt, J.G. 1994. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins, Baltimore, Maryland, USA. [ Links ]
Hu, S. & A.H.C. van Bruggen. 1997. Microbial dynamics associated with multiphasic decomposition of 14C-labeled cellulose in soil. Microb. Ecol. 33: 134-143. [ Links ]
Janssen, P.H. 2006. Identifying the Dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72: 1719-1728. [ Links ]
Jensen, G., B. Hansen, J. Eilenberg & J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5: 631-640. [ Links ]
Joshi, S.R., M. Chauhan, G.D. Sharma & R.R. Mishra. 1991. Effect of deforestation on microbes, vAM fungi and their enzymatic activity in Eastern Himalaya, p. 141-152. In G.S. Rajwas (ed.). Studies in Himalayan Ecobiology. Today and Tommorows Publication, New Delhi, India. Jost, L. 2006. Entropy and diversity. Oikos 113: 363-375. [ Links ]
Kapur, M. & R.K. Jain. 2004. Microbial diversity: exploring the unexplored also available on-line: http:// crdd.net/open/962/1/Jain2004.1.pdf. [ Links ]
Kier, G. & W. Barthlott. 2001. Measuring and mapping endemism and species richness: a new methodological approach and its application on the flora of Africa. Biodivers. Conserv. 10: 1513-1529. [ Links ]
Kirk, J.L., L.A. Beaudette, M. Hart, P. Moutoglis, J.N. Klironomos, H. Lee & J.T. Trevors. 2004. Methods of studying soil microbial diversity. J. Microbiol. Methods 58: 169-188. [ Links ]
Kowalchuk, G.A., D.S. Buma & W. De Boer. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Leeuwenhoek 81: 209-520. [ Links ]
Krebs, C.J. 1978. Ecology: The Experimental analysis of Distribution and Abundance. Harper and Row, New York, New York, USA. [ Links ]
Kumar, S. 2011. Fluctuation of soil bacterial dehydrogenase activity in response to the application of Endosulfan and Chlorpyrifos. J. Cell Tissue Res. 11: 2847-2851. [ Links ]
Kumar, R., C. Acharya & S.R. Joshi. 2011. Isolation and analyses of uranium tolerant Serratia marcescens strains and their utilization for aerobic uranium U(vI) bioadsorption. J. Microbiol. 49: 568-574. [ Links ]
Lee, C.S., Y.T. Jung, S. Park, T.K. Oh & J.H. Yoon. 2010. Lysinibacillus xylanilyticus sp. nov., a xylan-degrading bacterium isolated from forest humus. Int. J. Syst. Evol. Microbiol. 60: 281-286. [ Links ]
Lin, Y.T., Y.J. Huang, S.L. Tang, W.B. Whitman, D.C. Coleman & C.Y. Chiu. 2010. Bacterial community diversity in undisturbed perhumid Montane forest soils in Taiwan. Microb. Ecol. 59: 369-378. [ Links ]
Lomolino, M.v. 2001. Elevation gradients of species density: historical and prospective views. Glob. Ecol. Biogeogr. 10: 3-13. [ Links ]
Lovett, J.C., S. Rudd, J. Taplin & C. Frimodt-Moller. 2000. Patterns of plant diversity in Africa south of the Sahara and their implications for conservation management. Biodivers. Conserv. 9: 37-46. [ Links ]
Magurran, A.E. 2004. Measuring biological diversity.Blackwell, Oxford, United Kingdom. [ Links ]
Makoi, J.H.J.R. & P.A. Ndakidemi. 2008. Selected soil enzymes: Examples of their potential roles in the ecosystem. Afr. J. Biotechnol. 7: 181-191. [ Links ]
May, R.M. 1975. Patterns of species abundance and diversity, p. 81-120. In M.L. Cody & J.M. Diamond (eds.). Ecology and Evolution of Communities. Harvard University, Cambridge, Massachusetts, USA. [ Links ]
Moore, A.W. & J.S. Russell. 1972. Factors affecting dehydrogenase activity as an index of soil fertility. Plant Soil 37: 675-82. [ Links ]
Myers, N., R.A. Muttermeier, C.A. Muttermeier, G.A.B. DaFornseca & J. Kent. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853-858. [ Links ]
Palmer, C., E.M. Bik, M.B. Eisen, P.B. Eckburg, T.R. Sana, P.K. Wolber, D.A. Relman & P.O. Brown. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34: e5. [ Links ]
Pandey, A., P. Trivedi, B. Kumar, B. Chaurasia & L.M.S. Palni. 2006. Soil microbial diversity from the Himalaya: Need for documentation and conservation. p. 64, vol. 5, NBA Scientific Bulletin, Chennai, India. [ Links ]
Paul, E.A. & A.D. Mclaren. 1975. Biochemistry of the soil subsystem, p. 1-36. In E.A. Paul & A.D. McLaren (eds.). Soil Biochemistry. Marcel Dekker, New York, USA. [ Links ]
Pielou, E.C. 1966. The measurement of diversity in different types of biological collections. J. Theor. Biol. 13: 131-144. [ Links ]
Rondon, M.R., M. Robert, G. Handelsman & J. Handelsman. 1999. The Earth’s bounty: assessing and accessing soil microbial diversity. Trends Biotechnol. 17: 403-409. [ Links ]
Shivaji, S., P. Chaturvedi, Z. Begum, P.K. Pin, R. Manorama, D.A. Padmanaban, Y.S. Shouche, S. Pawar, P. vaishampayan, C.B.S. Dutt, G.N.R. Datta, K. Manchanda, U.R. Rao, P.M. Bhargava & J.v. Narlikar. 2009. Janibacter hoylei sp. nov., Bacillus isronensis sp. nov. and Bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int. J. Syst. Evol. Microbiol. 59: 2977-2986. [ Links ]
Subhani, A., H. Changyong, x. Zhengmiao, L. Min & M. El-ghamy. 2001. Impact of soil environment and agronomic practices on microbial/dehydrogenase enzyme activity in soil. A review. Pakistan J. Biol. Sci. 4: 333-338. [ Links ]
Sumathi, T., A. Janardhan, A. Srilakhmi, D.v.R. Sai Gopal & G. Narasimha. 2012. Impact of indigenous microorganisms on soil microbial and enzyme activities. Arch. Appl. Sci. Res. 4: 1065-1073. [ Links ]
Tabatabai, M.A. 1977. Effect of trace elements on urease activity in soils. Soil Biol. Biochem. 9: 9-13. [ Links ]
Tabatabai, M.A. 1994. Soil enzymes, p. 797-798. In R.W. Weaver, J.R. Angle & P.S. Bottomley (eds.). Methods of Soil Analysis, part 2. Soil Science Society of America, Madison, USA. [ Links ]
Teixeira, L.C.R.S., R.S. Peixoto, J.C. Cury, W.J. Sul, v.H. Pellizari, J. Tiedje & A.S. Rosado. 2010. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. The ISME Journal 4: 989-1001. [ Links ]
Thomson, B.C., N. Ostle, N. McNamara, M.J. Bailey, A.S. Whiteley & R.I. Griffiths. 2010. vegetation affects the relative abundances of dominant soil bacteria taxa and soil respiration rates in an upland grassland soil. Microb. Ecol. 59: 335-343. [ Links ]
Vilain, S., Y. Luo, M. Hildreth & v. Brozel. 2006. Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl. Environ. Microbiol. 72: 4970-4977. [ Links ]
Vishniac, H.S. 1993. The microbiology of Antarctic soils, p. 297-341. In E.I. Friedmann (ed.). Antarctic microbiology. Wiley-Liss, New York, USA. Ward, B.B. 2002. How many species of prokaryotes are there. PNAS 99: 10234-10236. [ Links ]
Wick, B., R.F. Kühne & P.L.G. vlek. 1998. Soil microbiological parameters as indicators of soil quality under improved fallow management systems in southwestern Nigeria. Plant Soil 202: 97-107. [ Links ]
Yadav, S., R. Kaushik, K.A. Saxena & K.D. Arora. 2011. Genetic and functional diversity of Bacillus strains in the soils long-term irrigated with paper and pulp mill effluent. J. Gen. Appl. Microbiol. 57: 183 195. [ Links ]
Zak, J.C., M.R. Willig, D.L. Moorhead & H.G. Wildman. 1994. Functional diversity of microbial communities: a quantitative approach. Soil Biol. Biochem. 26: 1101-1108. [ Links ]
Zar, J.H. 1984. Biostatistical Analysis. Prentice Hall, Upper Saddle River, New Jersey, USA. [ Links ]
Zaved, H.K., M. Mizanur Rahman, M. Mashiar Rahman, A. Rahman, S.M.Y. Arafat & M. Safiur Rahman. 2008. Isolation and characterization of effective bacteria for solid waste degradation for organic manure. KMITL Sci. Tech. J. 8: 44-55. [ Links ]
*Correspondencia a:
Nathaniel A. Lyngwi. Microbiology Laboratory, Department of Biotechnology & Bioinformatics North-Eastern Hill University, Shillong Meghalaya, India; nlyngwi@gmail.com.
Khedarani Koijam. Microbiology Laboratory, Department of Biotechnology & Bioinformatics North-Eastern Hill University, Shillong Meghalaya, India; khedarani_k@yahoo.com.
D. Sharma. Research Officer, Regional Centre-NAEB, North-Eastern Hill University, Shillong, Meghalaya, India. dhrubasharma07@gmail.com
S. R. Joshi. Microbiology Laboratory, Department of Biotechnology & Bioinformatics North-Eastern Hill University, Shillong Meghalaya, India; srjoshi2006@yahoo.co.in.
1. Microbiology Laboratory, Department of Biotechnology & Bioinformatics North-Eastern Hill University, Shillong Meghalaya, India; srjoshi2006@yahoo.co.in, khedarani_k@yahoo.com, nlyngwi@gmail.com, dhrubasharma07@gmail.com
2. Research Officer, Regional Centre-NAEB, North-Eastern Hill University, Shillong, Meghalaya, India.
Received 27-II-2012. Corrected 10-VIII-2012. Accepted 19-IX-2012.