Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.59 n.3 San José Sep. 2011
Characterization of Bacillus thuringiensis soil isolates from Cuba, with insecticidal activity against mosquitoes
Aileen González1, Raúl Díaz2, Manuel Díaz1, Yainais Borrero3, Rosa Y. Bruzón1, Bertha Carreras4 & René Gato1
1. Department of Vector Control. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; aileen@ipk.sld.cu, mdiazp@ipk.sld.cu, yirian@ipk.sld.cu, rene@ipk.sld.cu
2. Department of Bacteriology-Mycology. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; raul.diaz@ipk.sld.cu
3. Department of Diagnostic Laboratory. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; yaini@ipk.sld.cu
4. INISAV Research Institute of Plant Protection. Havana, Cuba; bcarreras@inisav.cu
1. Department of Vector Control. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; aileen@ipk.sld.cu, mdiazp@ipk.sld.cu, yirian@ipk.sld.cu, rene@ipk.sld.cu
2. Department of Bacteriology-Mycology. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; raul.diaz@ipk.sld.cu
3. Department of Diagnostic Laboratory. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; yaini@ipk.sld.cu
4. INISAV Research Institute of Plant Protection. Havana, Cuba; bcarreras@inisav.cu
Dirección para correspondencia
Abstract
Chemical insecticides may be toxic and cause environmental degradation. Consequently, biological control for insects represents an alternative with low ecological impact. In this work, three soil isolates (A21, A51 and C17) from different regions of the Cuban archipelago were identified, characterized and evaluated against Aedes aegypti and Culex quinquefasciatus. The new isolates were compared with reference IPS82 strain and two strains isolated from biolarvicides Bactivec and Bactoculicida, respectively. The differentiation was done by morphological, biochemical, bioassays activity and molecular methods (SDS-PAGE, plasmid profile and random amplified polymorphic analysis). All isolates were identified as Bacillus thuringiensis. The A21, A51 and C17 isolates showed higher larvicide activity than Bactivec’s isolated reference strain, against both A. aegypti and C. quinquefasciatus. A21 isolate had a protein profile similar to IPS82 and Bactivec strain. A51 and C17 isolates produced a characteristic proteins pattern. A21 and A51 isolates had plasmid patterns similar to IPS82 standard strain, while C17 isolate had different both plasmid profile and protein bands. All the studied isolates showed a diverse RAPD patterns and were different from the strains previously used in biological control in Cuba. Rev. Biol. Trop. 59 (3): 1007-1016. Epub 2011 September 01.
Key words: Bacillus thuringiensis, biological control, entomopathogenic bacteria, Aedes aegypti, Culex quinquefasciatus, SDS-PAGE, plasmid, RAPD
Resumen
El uso prolongado de insecticidas ha conducido al desarrollo de resistencia en diferentes especies de mosquitos y al incremento de la degradación del ambiente. El control biológico de insectos ha devenido como una alternativa útil y de bajo impacto ambiental. En nuestro estudio fueron identificados, caracterizados tres aislamientos de suelos procedentes de diferentes regiones del archipiélago cubano y comparados con cepas de referencia: aisladas de los biolarvicidas Bactivec y Bactoculicida, además de IPS82. La diferenciación de los mismos se llevó a cabo mediante métodos morfológicos, bioquímicos y moleculares (SDSPAGE, perfil plasmídico, RAPD). Los aislamientos fueron identificados como Bacillus thuringiensis; A21, A51 y C17 mostraron una mayor actividad contra larvas de Aedes aegypti and Culex quinquefasciatus que la cepa aislada del biolarvicida Bactivec, utilizada como referencia en este estudio. Dos de los aislamientos poseían perfiles proteicos y plasmídicos similares al de la cepa control IPS82, pero el restante difería de ellos. Los tres mostraron patrones de RAPD diferentes lo que nos permitió su diferenciación. Estos patrones de RAPD también diferían del observado para las cepas utilizadas comúnmente en el control biológico en nuestro país.
Palabras claves: Bacillus thuringiensis, control biológico, bacterias entomopatógenas, Aedes aegypti, Culex quinquefasciatus, SDS-PAGE, plásmidos, RAPD.
Vector-borne diseases are one of the major public health problems in developing countries. Aedes aegypti (A. aegypti) (Diptera: Culicidae) is the main vector of Dengue fever and Culex quinquefasciatus (C. quinquefasciatus) (Diptera: Culicidae) could be vector of filariasis and West Nile encephalitis. Since vaccines are not available, the best control methods for these diseases are based on vector control. Chemicals insecticide applications had led to the emergence and spread of resistance in vectors of human diseases and to environmental degradation (Ben-Dov et al. 1997, Ibarra et al. 2003). Consequently, the biological control has been accepted as an alternative as part of the integrate pest management, with a low environmental impact (Nunes & Fiuza 2003). The use of entomopathogenic bacteria like Bacillus thuringiensis as larvicides is a viable alternative for insect control. Water-dispersible granules based on Bacillus thuringiensis serovar. israelensis (Bti) has been recommended by World Health Organization (WHO) for mosquito larvae control in container habitats (WHO 2009).
B. thuringiensis strains have showed a wide range of specificity for insects like Lepidopteran, Coleoptera, Hymenoptera, Homoptera, Mallophaga and Diptera. Intensive screening programs have isolated and characterized B. thuringiensis from soil samples, plant surface, dead insects, stored grains and aquatic habitats (Bravo et al. 1998, Reyes- Ramirez & Ibarra 2005, Reyes-Ramirez & Ibarra 2008).
The entomopathogenic activity of this bacterium is principally due to the presence of proteinaceous inclusions known as insecticidal crystal proteins (Cry proteins). These Cry proteins are coded by cry genes harbored in megaplasmid and they exhibit a variety of biological actions, including cytolitic, haemolytic action, in addition to entomocidal activities (Lima et al. 2002).
The detection and identification of wild Bacillus spp. isolates may be of great value due to highly specific action and with no effect on other non- target insects, plants and domestic animals (Lima et al. 2002). We identified, characterized and evaluated the insecticidal activity of Bacillus environmental isolates from different regions of the Cuban archipelago.
Materials and methods
Strains: Reference strains used in the study are listed as below: IPS 82 from the International Entomopathogenic Bacillus Centre, Institute Pasteur, Paris, France (B. thuringiensis serotype H-14). Strains isolated from most extensive biolarvicides used in Cuba: Bactivec (Labiofam, Cuba; B. thuringiensis serotype H-14) and Bactoculicida (powder formulation from the former Soviet Union, applied in Cuba in eighteen decade).
Native isolates: Soil samples from different regions of Cuba were collected in glass flasks. About 1g of soil was cultured in 10mL of Nutrient Broth for 24h at 30°C with orbital shaking at 150rpm. From the culture, 2mL was taken and heat-shocked in water bath at 60°C for 1h. Then the samples were spread on Nutrient Agar plates, incubated at 30°C for 24h and checked for the presence characteristic colonies of B. thuringiensis. Qualitative evaluation of larvicidal activity against A. aegypti was performed in order to select the most attractive isolates. Three isolates were taken from: A51, Cayo Largo del Sur (198°819’72” N-444°740’29” W); C17, Cayo Iguana (201°758’13” N-441°467’14” W) and A21, Santiago de Cuba city (171°813’32” N-608°837’76” W).
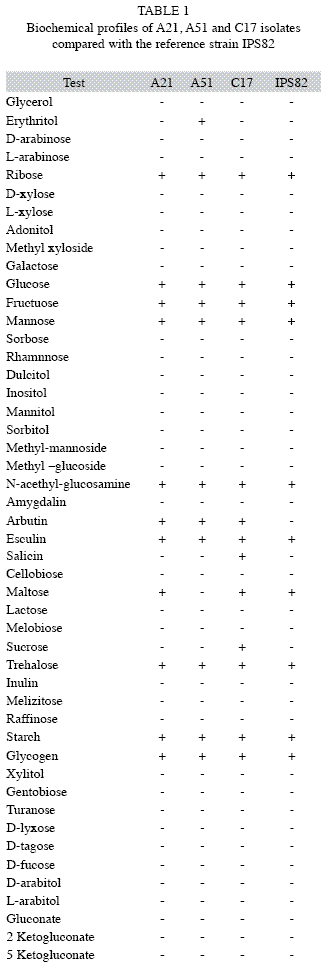
Biochemical and morphological characterization: The microbiological identification of the isolates was done by: Gram, lecithinase and haemolytic activity, screening for presence of parasporal bodies by phase-contrast microscopy. Biochemical profile was determined by API identification system 50 CHB (Biomerieux, France).
Bioassays: Preparation of active biomass (formulations): The isolates and Bactivec strain were grown in a fermentation medium based on sucrose, bacteriological peptone, yeast extract and inorganic salts (MgSO4 12.5mM; MnSO4 0.05mM; FeSO4 1.2 mM; ZnSO4 1.2mM; CaCl2 25mM) at 30°C 48-72h, until the formation of large amounts of spores and parasporal bodies. The World Health Organization doseresponse assay protocol was used to evaluate the susceptibilities of A. aegypti and C. quinquefasciatus to liquid formulations based on A21, A51 and C17 isolates (WHO 1999). The bioassays were undertaken by placing groups of 25 late 3rd instar larvae in 100mL of distilled water in 125mL plastic cups with the desired concentration of the bacterial formulations. At least five concentrations giving mortality between 10 and 90% were tested and mortality was recorded at 24h. Four replicated per dose were performed. Bioassays were replicated on at least three different weeks. Fifty lethal concentrations (LC50) was determined using Log-probit analysis with a program indicating means and standard error (SE). The LC50 were expressed in mg/L. A strain isolated from biolarvicide Bactivec was used as reference for comparing the results of bioassays.
Protein electrophoresis (SDS-PAGE): The isolates were grown in nutrient broth (BioCen, Cuba) at 30°C ± 1°C under 175rpm agitation for 48-72h or until cellular lysis with releases spores and δ-endotoxin crystals. The suspensions were centrifuged (10 000g for 20min) and the pellets were washed twice with NaCl 1M and distilled water. The pellet was resuspended in 100µL of distilled water and 100µL of sample buffer (Tris-HCl 500 mmol/L pH 6.8, 10% SDS, 4% 2-Mercaptoethanol, 8% Glycerol, 0.1% Bromophenol blue) and boiled at 100°C for 6min.
The protein profiles of the crystal components were determined through Sodium Dodecyl Sulfate (SDS)-Poliacrilamide gel electrophoresis (PAGE) (Laemmli & Favre 1973) with 10% Acrylamide separating gels. Ten µL of each sample were loaded onto gel immediately before electrophoresis.
Plasmid DNA analysis: Each strain was cultured in 50mL Luria-Bertani (LB) broth (Merck), to an optical density at 600nm of 0.9 to 1.1 at 30°C and 250rpm shaking. Vegetative cells were centrifuged at 20 200g for 15 min at 4°C. Each pellet was resuspended in 20mL cold TES buffer (30mM Tris base, 5mM EDTA, 50mM NaCl; pH 8.0) and centrifuged under the same conditions. Cells were resuspended in 2mL lysis buffer (TES buffer containing 20% Sucrose, 2mg/mL Lysozyme, and 1µL/ mL of RNase from a 10mg/mL stock solution) and incubated at 37°C for 3h. The spheroplast suspension was supplemented with 3mL of 8% SDS in TES buffer and incubated at 68°C for 10min. Then, 1.5mL of 3M Sodium acetate (pH 4.8) was added, and the suspension was incubated at -20°C for 30min. The suspension was centrifuged at 20 200g for 20min at 4°C. The supernatant was translucent; if it was not, another centrifugation was done, and ultimately, if still required, it was filtered. Two volumes of cold absolute ethanol were added to the supernatant and incubated overnight at -20°C. Plasmid-enriched DNA was centrifuged at 20 200g for 20min at 4°C. Each pellet was dissolved in 100µL Tris-EDTA (pH 8.0) (10mM Tris-HCl, 1mM EDTA) and stored at -20°C until further use.
In order to visualize the plasmid pattern from each strain, 10µL of each plasmidenriched DNA solution was loaded, along with the analytical marker DNA wide range (Promega), in 0.5% agarose gels (12 by 15cm) with 1x Tris-borate-EDTA buffer (45mM Tris-borate, 1mM EDTA), Ethidium bromide at 0.5µg/mL and run at 2V/cm for 20h. All gels included the plasmid preparation from IPS82 strain as a reference and strains isolated from biolarvicides Bactivec and Bactoculicida (Reyes- Ramírez & Ibarra 2008).
Random amplified polymorphic DNA (RAPD) analysis: Bacterial cells were grown overnight in 5mL of LB broth at 28°C with shaking. One mL of the culture was transferred to another tube with 2mL of Nuclisens lysis buffer. DNA extraction was performed following the instructions for Nuclisens Magnetic extraction (Biomerieux, France).
RAPD test was performed using nine primers (8, 10, 11, 12, 14, 15, 16, 17 and 18) from OPA Kit A (Operon Technologies, USA). Each polymerase chain reaction (PCR) reaction (20µL final volume) contained 1µL of DNA, 0.25pmoles of each primer, 10mmol/L of dNTPs, 1.5mmol/L MgCl2 and 1U Taq DNA polymerase (Promega, USA). DNA amplification reactions were performed in a DNA thermocycler PCT-150 (MJ Research, Inc, USA) programmed as follows: 2 min at 95°C, followed by 35 cycles composed of 30 s at 95°C, 1 min at 30°C, 2 min at 72°C and a final extension of 5 min at 72°C.
Amplified product were analyzed in 1% agarose gel in Tris Borate EDTA (TBE) with ethidium bromide and compared with the molecular markers λHindIII (Promega, USA) and Gene Ruler 100 pb DNA ladder plus (Fermentas, Lithuania).
Each band produced in the RAPD reactions were considered as an individual character (present or absent).
The results of API identification system 50 CHB and RAPD analysis were transformed into binary matrixes (1/0) and evaluated with the bioinformatic program Multivariate statistic package (MVSP 3.1) (Kovach company services, UK). Profile analysis was performed based on the calculation of the matching similarity coefficient (Jaccard coefficient), used to establish the similarity matrix. This matrix was transformed into a phenogram using the unweighted pair group method (UPGMA).
Results
Biochemical and morphological characterization: The three isolates were Gram positive rods, lecithinase positive, showed haemolytic activity in blood agar and produced spores in oval way. Biochemical test results of A21, A51, C17 isolates and the reference strain IPS 82 are shown in Table 1. Based on the biochemical characteristics, the A21, A51 and C17 isolates had compatible patterns with Bacillus cereus. Morphological observation under phase-contrast microscopy showed a single ovoid parasporal crystal, so the three strains were identified as B. thuringiensis. A21, A51 and C17 isolates showed a similarity coefficient to 0.90, 0.83 and 0.76, respectively, in relation to the reference strain.
Bioassays: Summary of data obtained from the series of bioassays using a WHO protocol (WHO 1999) is shown in Table 2. All the native isolates showed a better CL50 against both A. aegypti and C. quinquefasciatus than reference strain (Bactivec).
SDS-PAGE analysis: The SDS-PAGE of spore-crystal suspensions of selected isolates is shown in Fig. 1. Each isolates produced a characteristic band pattern easily identified by the position of the major protein bands. The patterns were reproducible for the replicates of the strains in different gels. A21 had a protein profile similar to that reported for IPS82 (Ibarra et al. 2003) and Bactivec strain, with major proteins of 137, 70, 30, 28 and 18kDa. In the parasporal inclusions of A51 isolates a number of polypeptides with molecular masses of 18-137kDa were observed. C17 had different protein bands of 100, 85, 75, 52, 26 and 18kDa, with a major band to 26kDa. The electrophoretic gel patterns of A51 and C17 isolates were different to IPS82, Bactivec and Bactoculicida strains.
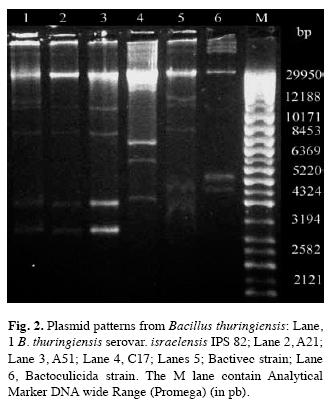
Plasmid profile: Fig. 2 show the plasmid profile of all B. thuringiensis strains included in this report. All strains had more than one plasmid. Megaplasmids were observed over chromosomal DNA. However, their poor migration into agarose gel made difficult the discrimination. But the plasmid migrating under chromosomal DNA allows us the comparison between the strains. A21 and A51 showed the same plasmid patterns to IPS82, but different to Bactivec and Bactoculicida strains. Moreover, C17 had a distinctive plasmid pattern.
RAPD analysis: The used of OPA primers produced a range of 2 to 11 bands, allowing to conform at least four different groups per primer. RAPD reactions with OPA 8 and OPA 12 primers yielded characteristic products of 4-6 bands ranging from 180 to 1800pb. In both cases, four different patterns were observed. Besides with OPA 10, 11, 14, 15, 16 and 17 primers five RAPD patterns were found. In contrast, OPA 18 primers exhibited six different patterns (Fig. 3). In general, the OPA 8, 11, 12, 16, 17 and 18 primers showed a better discriminatory power (Fig. 3).
A21 and A51 isolates showed the same electrophoretic patterns of IPS82 strain with OPA 8 and OPA 12 primers, but C17 isolate revealed a genetic distance of 0.22 with OPA 8 and 0.042 with OPA 12. On the other hand, A21 isolate and IPS82 showed identical RAPD pattern with the primers OPA 11 and 17, but A51 isolate varied the genetic distance from 0.90 (with OPA 11 primer) to 0.536 (with OPA 17 primer). The C17 isolate stayed distant of the control with the former primers (0.13 with OPA 11 and 0.35 with OPA 17). The obtained results with remain primers (OPA 10, 14, 15, 16 and 18) showed that A21 and A51 isolates increased the genetic distance in relation to the control. However, C17 isolate maintained a larger distance from them. With all used primers the three isolates were genetically distant from the biolarvicides Bactivec and Bactoculicida.
The complete RAPD patterns were used to create the first dendrogram, where A21 and A51 isolates showed a similarity of 0.76 and 0.73 with the control strain. C17 isolate had genetic differences with the other isolates (Data no shown).
The second dendrogram was built based on fragments generated by OPA 8, 11, 12, 16, 17 and 18 primers and suggested that A21 isolate is closer than IPS82. A51 isolate and IPS82 strain showed a 0.74 of similarity. On the contrary, C17 isolate results emphasize its dissimilarity with the other isolates and strains (Fig. 4).
Discussion
The characterization of microorganisms is essential for microbial ecology studies. Classification, identification and differentiation of microbes have traditionally relied on test based on phenotypic characteristics (Porcar et al. 1999). The phenotypic methods used for classifying B. thuringiensis, H-serotyping and biochemical characters, have both contributed to the establishment of an useful classification (Iriarte et al. 2000, Khyami-Horani et al. 2003).
Here we report the identification and characterization of three Cuban Bacillus isolates with larvicidal activity against A. aegypti and C. quinquefasciatus. There are previous studies in Cuba about native Bacillus isolates, but focused on agricultural pest management (Carreras & Piedra 2009).
We have demonstrated that A21, A51 and C17 isolates share most of the phenotypic features of B. thuringiensis strains previously classified (Lecadet et al. 1999, Porcar et al. 1999, Iriarte et al. 2000). The numerical analysis of biochemical patterns showed that A21 and A51 were closer related to reference strain than C17. The biochemical pattern compatible with B. cereus (by API 50 CH) and the presence of ovoid parasporal inclusion, allow us to classify the isolates as B. thuringiensis. However, there are not enough evidences for differentiation of these isolates by these methods.
In the biological assays, the isolates showed more efficacies against A. aegypti and C. quinquefasciatus than the B. thuringiensis strain currently used in a program of mosquito control in Cuba (biolarvicide Bactivec) (Table 2). A51 isolate had a better entomopathogenic activity of all strains. Interestingly, C17 had poor activity against A. aegypti; the LC50 against C. quinquefasciatus was eleven times higher than against A. aegypti.
These data are in agreement with the expected protein composition observed in the SDS-PAGE analyses. A21 isolate showed a high similarity with the control strain IPS82 and Bactivec, by mean of mosquitocidal protein profile. This isolate showed proteins common in B. thuringiensis serovar israelensis strains (Porcar et al. 1999, Ibarra et al. 2003, Sreshty et al. 2010).
Analysis by protein profile of A51 revealed the presence of polypeptides with molecular masses of 18 to 137kDa. The 28kDa polypeptide is produced in low levels poorly visualized in SDS-PAGE compared with the control IPS82 and Bactivec. The proteins from C17 were different from A51 and A21; three of them (100, 75 and 26kDa) have been described as active against mosquitoes (Ibarra et al. 2003).
The C17 isolate also had singular plasmid profile. On the other hand, the size of the plasmidic bands of A21 and A51 isolates were in the range described for other B. thuringiensis serovar israelensis strains previously analyzed (González & Carlton 1980, Porcar et al. 1999, Reyes-Ramirez & Ibarra 2008).
Molecular methods have emerged as the most rapid reliable and simple alternatives to characterize and differentiate microorganisms (Rademarker et al. 2005).
Genotyping of bacteria using PCR based methods, such as RAPD analysis is a commonly used approach for strain typing because it is economic, fast and simple (Malkawi et al. 1999, Romero & Orduz 2002, Rivera & Priest 2003, Vilas-Bôas & Lemos 2004, Arango et al. 2002). These methods also allow distinguishing isolates from commercial strains of B. thuringiensis (Brousseau et al. 1993, Fredireksen et al. 2006).
The combination of classical and molecular methods helps us to group the isolates and give more details than a single classification method. The bioinformatic analysis of the RAPD fingerprints using the most discriminatory primers helped to differentiate the native isolates from the used control strains.
We found that A21 was closer related to IPS82 strain than A51 and C17 isolates. Interestingly, the most genetically related isolates (A21 and A51) were obtained from distant geographical regions of Cuban archipelago (617km). Both, Santiago de Cuba and Canarreos archipelago have similar weather conditions and share most of the mosquito’s species, including C. quinquefasciatus (González 2006).
In contrast, C17 and A51 were isolated from close geographical keys of the same archipelago (separated by 4.4km) but showed great phenotypic and genetic differences. Although these small keys had been treated for years with B. thuringiensis for larvicide, it was demonstrated that all isolates were different from the commercial strains present in the microbial larvicides used in Cuba.
In conclusion, the new isolates showed a strong activity against A. aegypti and C. quinquefasciatus. Due to their distinctive characteristics C17 and A51 isolates are great candidates for the development of new larvicide formulations. The isolation of native strains with activity against pests, gives us new tools to be introduced into the mosquito management program. This will decrease the disturbance of ecosystem balance through reducing the release of chemical pesticides or other exogenous biological products.
References
Arango, J.A., M. Romero & S. Orduz. 2002. Diversity of Bacillus thuringiensis strains from Colombia with insecticidal activity against Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Appl. Microbiol. 92: 466-474. [ Links ]
Ben-Dov, E., A. Zaritsky, E. Dahan, Z. Barak, R. Sinai, R. Manasherob, A. Khamraev, E. Troistskaya, A. Dubitsky, N. Berezina & Y. Margalith. 1997. Extended screening by PCR for seven Cry-group genes from field-collected strains of Bacillus thuringiensis. Appl. Environ. Microbiol. 63: 4883-4890. [ Links ]
Bravo, A., S. Sarabia, L. López, H. Ontiveros, C. Abarca, A. Ortiz, M. Ortiz, L. Lina, F. Villalobos, G. Peña, M. Nuñez-Valdez, M. Soberon & R. Quintero. 1998. Characterization of cry genes in Mexican B. thuringiensis strain collection. Appl. Environ. Microbiol. 64: 4965-4972. [ Links ]
Brousseau, R., A. Saint-Onge, G. Préfontaine, L. Masson & J. Cabana. 1993. Arbitrary primer polymerase chain reaction a powerful method to identify Bacillus thuringiensis serovars and strains. Appl. Environ. Microbiol. 59: 114-119. [ Links ]
Carreras, B., D. Rodríguez & F. Piedra. 2009. Evaluación de cepas nativas de Bacillus thuringiensis Berliner para el control de Heliothis virescens Fabricius en el cultivo del tabaco en Cuba. Fitosanidad 13: 277-280. [ Links ]
Chaves, J.Q., C.F. Cavados & L. Rabinovitch. 2008. Phenotypic and genotypic features of a new autoagglutinating B. thuringiensis strains. J. Invert. Pathol. 98: 85-92. [ Links ]
Frederiksen, K., H. Rosenquist, K. Jorgensen & A. Wilcks. 2006. Occurrence of natural Bacillus thuringiensis contaminants and residues of Bacillus thuringiensisbased insecticides on fresh fruits and vegetables. Appl. Environ. Microbiol. 72: 3435-3440. [ Links ]
González, I. & A. Carlton. 1980. Patterns of plasmid DNA in crystalliferous and acrystalliferous strains of B. thuringiensis. Plasmid 3: 92-98. [ Links ]
González, R. 2006. Culícidos de Cuba. Editorial Científico Técnica. La Habana, Cuba. [ Links ]
Ibarra, J.E., M.C. del Rincón, S. Ordúz, D. Noriega, G. Benintende, R. Monnerat, L. Regis, C.M.F. de Oliveira, H. Lanz, M.H. Rodriguez, J. Sánchez, G. Peña & A. Bravo. 2003. Diversity of Bacillus thuringiensis strains from Latin America with insecticidal activity against different mosquito species. Appl. Environ. Microbiol. 69: 5269-5274. [ Links ]
Iriarte, J., V.C. Dumanoir, Y. Bel, M. Porcar, M.D. Ferrandis, M.M. Lecadet, J. Ferre & P. Caballero. 2000. Characterization of Bacillus thuringiensis ser. balearica (Serotype H48) and ser. navarrensis (Serotype H50): two novel serovars isolated in Spain. Curr. Microbiol. 40: 17-22. [ Links ]
Khyami-Horani, H., M. Hajaij & C. Jean-Francois. 2003. Characterization of Bacillus thuringiensis ser. Jordanica (Serotype H71), a novel serovariety isolated in Jordan. Curr. Microbiol. 47: 26-31. [ Links ]
Laemmli, U.K. & M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80: 575-599. [ Links ]
Lecadet, M.M., E. Frachon, V. Cosmao, H. Ripouteau, S. Hamon, P. Laurent & I. Thiéry. 1999. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 86: 660-672. [ Links ]
Lima, A.S.G., A.M. Guidelli, I.L. Abreu & M.V.F. Lemos. 2002. Identification of new isolates of Bacillus thuringiensis using rep-PCR products and δ-endotoxine electron microscopy. Gen. Mol. Biol. 25: 225-229. [ Links ]
Malkawi, H.I., F. Al-Momani, M.M. Meqdom, I. Saadoun & M.J. Mohammad. 1999. Detection of genetic polymorphism by RAPD-PCR among isolates of B. thuringiensis. New Microbiol. 22: 241-247. [ Links ]
Nunes, L.M. & L.M. Fiuza. 2003. PCR and bioassays secreening of Bacillus thuringiensis isolates from rice-fields of Rio Grande Do Sul, specific to lepidopterans and coleopterans. Braz. J. Microbiol. 34: 305-310. [ Links ]
Porcar, M., J. Iriarte, V. Cosmao, M.D. Ferrandis, M.M Lecadet, J. Ferré & P. Caballero. 1999. Identification and characterization of the new B. thuringiensis serovars pirenaica (serotype H57) and iberica (serotype H59). J. Appl. Microbiol. 87: 640-648. [ Links ]
Rademarker, J.L., J.M. Aarts & P. Vinuesa. 2005. Molecular typing of environmental isolates, p. 97-126. In O. Mark & C. Smith (eds.). Molecular Microbiol Ecology. New York, New York, USA. [ Links ]
Reyes-Ramires, A. & J.E. Ibarra. 2005. Fingerprinting of Bacillus thuringiensis type strains and isolates by using Bacillus cereus group-specific repetitive extragenic palindromic sequence-based PCR analysis. Appl. Environ. Microbiol. 71: 1346-55. [ Links ]
Reyes-Ramírez, A. & J.E. Ibarra. 2008. Plasmid patterns of B. thuringiensis type atrains. Appl. Environ. Microbiol. 74: 125-129. [ Links ]
Rivera, A.M. & F.G. Priest. 2003. Molecular typing of B. thuringiensis serovars by RAPD-PCR. Syst. Appl. Microbiol. 26: 254-261. [ Links ]
Sreshty, M.A., S. Misra & U.S. Murty. 2010. Interpreting the SDS-PAGE protein patterns with self-organizing maps: application for the characterization of mosquito- pathogenic Bacillus strains. J. Appl. Microbiol. 110: 239-247. [ Links ]
World Health Organization. 2009. Dengue: guidelines for diagnosis, treatment, prevention and control. WHO. (Accessed: 9 November 2010, whqlibdoc.who.int/publications/2009/9789241547871_eng.pdf. [ Links ])
World Health Organization. 2009. Dengue: guidelines for diagnosis, treatment, prevention and control. WHO. (Accessed: 9 November 2010, whqlibdoc.who.int/publications/2009/9789241547871_eng.pdf) [ Links ]
Vilas-Bôas, G. & M. Lemos. 2004. Diversity of cry genes and genetic characterization of Bacillus thuringiensis isolated from Brazil. Can. J. Microbiol. 50: 605-613. [ Links ]
Correspondencia a: Aileen González, Manuel Díaz, Rosa Y. Bruzón & René Gato. Department of Vector Control. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; aileen@ipk.sld.cu, mdiazp@ipk.sld.cu, yirian@ipk.sld.cu, rene@ipk.sld.cu
Raúl Díaz. Department of Bacteriology-Mycology. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; raul.diaz@ipk.sld.cu
Yainais Borrero. Department of Diagnostic Laboratory. Institute of Tropical Medicine “Pedro Kourí”, Havana, Cuba. PO Box 601, Marianao 13, Ciudad de La Habana, Cuba; yaini@ipk.sld.cu
Bertha Carreras. INISAV Research Institute of Plant Protection. Havana, Cuba; bcarreras@inisav.cu
Received 17-VIII-2010. Corrected 04-I-2011. Accepted 27-I-2011.