Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.55 n.2 San José Jun. 2007
Characterization and phylogenetic relationships of strains of Dengue type 1 virus from Costa Rica
César-Alonso Cerdas-Quesada1* & Axel P. Retana-Salazar2
1 Caja Costarricense del Seguro Social, Hospital San Juan de Dios, Departamento de Análisis Clínico y Microbiología. San José, Costa Rica.
* Correspondence and reprint requests should be addressed at PO Box 10878-1000 San José, Costa Rica, Central America. Tel (506)8117406, Fax (506)2271109; cesar.virologia.ucr@costarricense.cr
2 Centro de Investigación en Estructuras Microscópicas, Ciudad de la Investigación, Universidad de Costa Rica, 2060, San José, Costa Rica.
Received 22-IV-2003. Corrected 12-II-2007. Accepted 19-II-2007.
Abstract: Phylogenetic analysis of the envelope gene sequence (E) of three isolated Dengue virus type 1 (DEN1) sequences from the Costa Rican 1993 epidemic was performed. All the strains fell into genotype V. Genotype V has grouped all the strains isolated from America, three strains from Africa and two strains from Asia. Costa Rican strains are closely related to the strain from Angola and to the Brazil 1990 strain. Costa Rican strains are able to cause DHF but with low clinical disease severity and low incidence maybe because of low virulence. Rev. Biol. Trop. 55 (2): 365-372. Epub 2007 Jun 29.
Key words: Dengue virus, phylogeny, molecular epidemiology, envelope, Flavivirus, Costa Rica.
The three genera of the Family Flaviviridae, i.e., Flavivirus, Pestivirus, and Hepatitis C virus, comprise enveloped viruses with single-straded plus-sense RNA genomes (Wengler et al. 1995, Lanciotti et al. 1997). The genus Flavivirus comprises important human pathogens as the Dengue (DEN) virus, Japanese encephalitis (JE) virus, St. Louis encephalitis (SLE) virus, West Nile (WN) virus, Tick-borne encephalitis (TBE) virus and the Yellow Fever (YF) virus (Wallner et al. 1998). They are transmitted between humans by insects and tick vectors and are currently endemic throughout most tropical areas of the world (Gubler 1998, Weaver et al. 1999).
DEN viruses occur as four antigenically distinct serotypes, which do not induce cross protection. Infection with any of those leads to a mild, self-limiting febrile illness (Dengue fever, DF) (Goncalvez et al. 2002). A secondary infection by a heterologous DEN serotype has been established as the main risk for the occurrence of a more severe form of the disease, Dengue hemorrhagic fever/ Dengue Shock Syndrome (DHF/DSS), which is respon sible for a high mortality rate, especially in children (Twiddy et al. 2002). Genetic variation has been studied with the four DEN serotypes, the analysis were based mainly on the comparisons of the sequences of genes coding for structural proteins (Rico Hesse 1990, Lewis et al. 1993, Lanciotti et al. 1995, Kuno et al. 1994, 1997). Dengue type 1 virus (DEN-1) was intro duced in America in 1997 and it has been responsible of several epidemics in different countries like Jamaica and Cuba, and Puerto Rico and Venezuela in 1978 (Pinheiro 1989, Pinheiro and Cobet 1997). In the succeeding 4 years, this serotype was spread throughout the Caribbean Islands, Mexico to Texas, Central America, and northern South America, causing major and minor epidemics. Prior to 1997, only DEN-2 and DEN-3 viruses were known to be present in the Americas, although DEN-1 was probably present in the early 1940s (Gubler and Meltzer 1999). The evolutionary rates of epidemic DEN-1 and DEN-2 have been seen similar, although the transmission pathways of these viruses around the world are different (Rico-Hesse 1990). Currently are recognized five genetic types of DEN-1: genotype I is represented by two early isolates from Hawaii, Southeast Asia, China and East Africa; genotype II, represented by strains isolated in Thailand; genotype III, represented by one sylvatic strain from Malaysia; genotype IV including isolates from West Pacific islands and Australia and genotype V, grouping strains isolated from America, West Africa and Asia (Goncalvez et al. 2002).
To date, there are not phylogenetic studies of DEN-1 in Costa Rica because of it we performed a phylogenetic analysis of DEN-1 to determinate the genogroup of the three isolated strains from Costa Rica what caused an epidemics in 1993.
Materials and methods.
Virus: DEN-1 strains sequenced in this article included three isolates from 1993 epidemics in Costa Rica consisting on the first passage in C6/36 cells of the original patients sera provided by Jeffry Chang (Centre for Control of Diseases, CDC, Atlanta, USA). All these viruses were isolated from human sera and were identified as DEN-1 serologically (Hemagglutination inhibition). Each sampled was isolated by a single passage in C6/36 cells. Viral RNA was extracted from C6/36 viral seed using Quiagen RNA extraction kit (Quiagen Inc, Valencia, CA) as the protocol provided by the manufacture. Titan reverse transcription (RT)-PCR kit was used to amplify E gene region sequence. The RT-PCR products were sequenced using both directions with dideoxy dye termination kit and sequence resolve using an ABI 377 automate sequencer. Sequences trace files analyzed, and sequence complied by using SeqMan Program of DNAStar software.
Nucleotide Sequences characterization and analysis: The nucleotide sequences of the E gene from three strains DEN-1 were determined Through the consensus sequence obtained for each virus presents after four separate determinations (GenBank accession numbers AY153755-AY153757). The E gene sequences of the Costa Rican isolates were compared with the homologous sequences of DEN-1 viruses isolated worldwide and were aligned with 31 sequences already reported to the GenBank. All the sequences were 1485 nucleotides long approximately and we observed the lack of hipervariability regions among strains.
Sequence analysis of DEN-1 strains: Phylogenetic analysis was performed using distance (Neighbour-joining, NJ) and parsimony (Heuristic Search with bootstrap and Jack-knife supports). The multiple sequence alignment was peformed with the program Clustal X (ver. 1.8, Thompson et al. 1997) and the program PAUP* (ver. 4b10) was used for all the phylogenetic analysis.
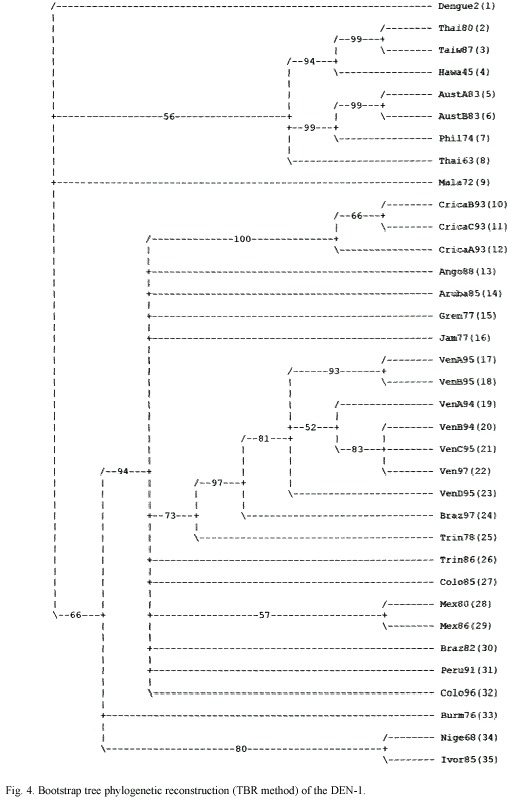
Unless stated otherwise, the following settings were used: Heuristic search, random addition with 100 replicates and unweighted characters; and, a NJ. Statistical support for clades was estimated by bootstrapping and Jack-knife with the fast procedure on 100 and 1000 replicates. Tree was rooted using homologous sequences of Japanese encephalitis (JE) virus, Kunjin (KUN) virus and Murray Valley Encephalitis virus (MVE).
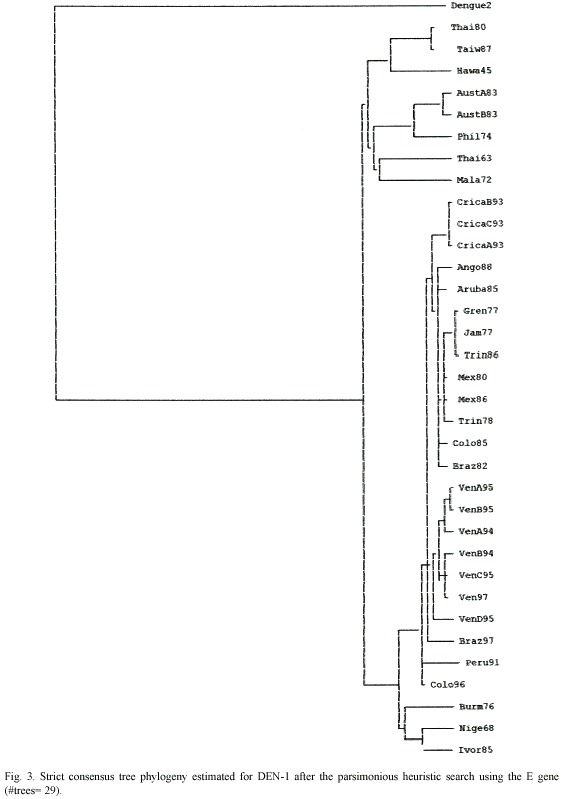
Trees were rooted using the homologous sequences of DEN-2 E gene previously deposited on GenBank data base. To determination of the genotype was performed by using parsimony (PAUP 4b10) method to obtain a strict bootstrap consensus.
Results
All the methods used produced trees with a topology of five genetic types/clades. A NJ tree of the four serotypes is presented in Fig. 1 and shows the Costa Rican strains as a group belonging to the Brazil90 lineage. The consensus tree of the DEN-1 genotypes is assigned to the Fig. 3 and 4 and shows that the Costa Rican isolates fall into the same genogroup, closely together (100% Jack-knife and bootstrap support; CI=0.7250; HI=0.2750; RI=0.7122) along with the other strain -Ango88- isolated from Angola collected in 1988. The sister group of the sequences from Costa Rica is formed by an Aruba isolate from 1985. The tree topology showed the five genetic types and it was supported by the bootstrap analysis and the Jack-knife 50% majority rule consensus tree that showed how the genotype V can be divided in three sub genotypes. The American isolates as the subgenotype VA, a 1976 Burma isolate as the sub genotype VB and, two isolates from Nigeria and Ivory Coast belonging to the sub genotype VC (Fig. 2)
Discussion
Epidemics attributed to DEN have been occurred at least in the last three centuries. DEN-1 has been circulated in several parts of the world in the first half of the 20th century. Its circulation has been demonstrated by serological evidence (Gubler 1997). Possibly a virus strain associated with a pandemic formed the ancestor lineage from which genotypes have been circulating in Asia, Oceania, and Africa in the last 50 years (Goncalvez et al. 2002).
Genetic diversity studies of DEN-1 has been analysed in previous studies. T1 fingerprint analysis allowed to the first classification in eight topotypes. Then, Rico-Hesse (1990) sequenced a fragment of 240 bases in E/NS1 junction and classified them in five genotypes. Goncalvez et al. 2002 produced evidence of the existence of five genotypes and the co-circulation of two different genotypes in America. Moreover, their results indicated that from the first introduction in 1977 only one type of DEN-1 has circulated in America with the potential to cause DF and DHF. Like most molecular studies of DEN, the main focus of the analysis is the E gene, which encodes the major protein component of the virion surface, is the most important antigen with regard to humoral immunity and is associated with other biological activities such as cell attachment, receptor binding and virus assembly (Twiddy et al. 2002).
Genotype V has grouped all the strains isolated from America, three strains from West Africa, and two strains from Asia. Costa Rican strains belong to genotype V and are closely related to the strain from Angola. Angola is the geographic origin proposed for the RIO H 36589 strain, since it was isolated from a Brazilian traveller who presented DF one week after his return to Brazil from Angola in 1988. Nevertheless, this strain was more closely related to the American than to the African lineage suggesting its American origin (Goncalvez et al. 2002). Costa Rican strains were closely related to a Brazil 1990 strain (Fig. 1). Costa Rican strains are able to cause DF and DHF, but with low clinical disease severity and DHF low incidence (Vornadam 2001). Venezuelan strains with potential to cause the most severe form of the disease belong to the same genotype (Goncalvez et al. 2002). Costa Rica has a few DHF/DSS cases maybe because of serotypes came at the same time or their low virulence. We are performing molecular evolutionary analysis of all serotypes genetic diversity in Costa Rica to determinate the processes shaping virus populations (Cerdas-Quesada and Retana-Salazar, unpublished).
Acknowledgments
We thank Jeffry Chang for laboratory advice and resources and for generously providing the DEN-1 sequences and Duane Gubler and Scott Weaver for providing relevant epidemiological information, as well as Federico Villalobos and Alejandro Valerio for comments of the manuscript.
Resumen
Tres aislamientos del gen de la envoltura (E) de Dengue tipo 1 del período epidémico de 1993 en Costa Rica se analizaron filogenéticamente. Todas las cepas pertenecen al genotipo V. El genotipo V ha agrupado todos los aislamientos de América, tres cepas de África y dos cepas de Asia. Los aislamientos costarricenses están cercanamente relacionados a la cepa de Angola y al aislamiento brasileño de 1990. Las cepas costarricenses son capaces de causar Fiebre de Dengue Hemorrágico pero con baja severidad clínica y baja incidencia quizá debido a una baja virulencia.
Palabras clave: Virus, Dengue, Flavivirus, filogenia, epidemiología molecular, Costa Rica.
References
Ashlock, P. 1974. The uses of cladistics. Ann. Rev. Ecol. System. 5: 81-99. [ Links ]
Goncalvez, A., A. Escalante, F. Pujol, J. Ludert, D. Tovar, R. Salas & F. Liprandi. 2002. Diversity and Evolution of the Envelope Gene of Dengue Virus Type 1. Virology 303: 110-119. [ Links ]
Gubler, D. 1998. Dengue and Dengue Hemorrhagic Fever. Clin. Microb. Rev. 11: 480-496. [ Links ]
Gubler, D. & M. Meltzer. 1999. Impact of Dengue/Dengue Hemorrhagic Fever on the Developing World. Advance. Virus Res. 53: 35-70. [ Links ]
Kuno, G., G. Chang, R. Tsuchiya, N. Karabatsos & B. Cropp. 1998. Phylogeny of genus Flavivirus. J. Virol. 72: 73-83. [ Links ]
Lanciotti, R., D. Gubler & D. Trent. 1997. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 78: 2279-2286. [ Links ]
Lewis, J., G. Chang, R. Lanciotti, R. Kinney, L Mayer & D. Trent. 1993. Phylogenetic Relationships of Dengue-2 Viruses. Virology 197: 216-227. [ Links ]
Pinheiro, F.P. 1989. Dengue in the Americas. 1980-1987. Epidemiol. Bull. 10: 1-8. [ Links ]
Pinheiro, F. & S.J. Corber. 1997. Global situation of dengue and dengue haemorrhagic fever and its emergence in the Americas. World Health Stat. Q. 50: 161-169. [ Links ]
Rico-Hesse, R. 1990. Molecular Evolution and Distribution of dengue Viruses Type 1 and 2 in Nature. Virology 174: 479-493. [ Links ]
Tolou, H., P. Couisisnier-Paris, J. Durand, V. Mercier, J. Pina, P. Micco, F. Billior & R. Charrel. 2001. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 82: 1283-1290. [ Links ]
Trent, D., J. Grant, T. Monath, C. Manske, M. Corina & G. Fox. 1989. Genetic Variation and Microevolution of dengue 2 Virus in Southeast Asia. Virology 172: 523-535. [ Links ]
Twiddy, S., J. Farrar, N. Vinh, B. Wills, E. Gould, T. Gritsun, G. Lloyd & E. Holmes. 2002. Phylogenetic relationships and Differential Selection Pressures among Genotypes of Dengue-2 Virus. Virology 298: 63-72. [ Links ]
Twiddy, S., C. Woelk & E. Holmes. 2002. Phylogentic evidence for adaptative evolution of dengue viruses in nature. J. Gen. Virol. 83: 1679-1689. [ Links ]
Uzcategui, N., D. Camacho, G. Comach, R. Cuello, E. Holmes & E. Gould. 2001. Molecular Epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J. Gen. Virol. 82: 2945-2953. [ Links ]
Vornadam, V. 2001. Dengue hemorrágico: Factores moleculares y climatológicos. El Dengue. Academia Nacional de Ciencias 7: 3-9 [ Links ]
Wallner, G., C. Mandl, C. Kunz & F. Heinz. 1995. The Flavivirus 3 Non-coding Region: Extensive Size Heterogeneity Independent of Evolutionary Relationships among strains of Tick-Borne Encephalitis Virus. Virology 213: 169-178. [ Links ]
Wang, E., H. Ni, R. Xu, A. Barrett, S. Watowich, D. Gubler & S. Weaver. 2000. Evolutionary Relationships of Endemic/Epidemic and Sylvatic Dengue Viruses. J. Gen. Virol. 74: 3227-3237. [ Links ]
Weaver, S., A. Broult, W. Kang, & J. Holland. 1999. Genetic Fitness Changes Accompanying Adaptation of an Arbovirus to Vertebrate and Invertebrate Cells. J. Virol. 73: 4316-4326. [ Links ]