Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de Biología Tropical
On-line version ISSN 0034-7744Print version ISSN 0034-7744
Rev. biol. trop vol.53 suppl.1 San José May. 2005
Development and field application of a molecular probe for the primary pathogen of the coral disease white plague type II
Laurie L.Richardson,DeEtta K.Mills,Elizabeth R.Remily &Joshua D.Voss.
Department of Biological Sciences,Florida International University,Miami,Florida 33199, USA; Laurie.Richardson@fiu.edu
Received 15-I-2004. Corrected 25-IX-2004. Accepted 29-III-2005.
Abstract:One of the current problems in the field of coral disease research is that of tracking coral pathogens in the natural environment.A promising method to do this is by use of pathogen-specific molecular probes. However,this approach has been little used to date.We constructed,and validated in the laboratory,a fluoro-chrome-labeled molecular probe specific to Aurantimonas coralicida ,the bacterial pathogen of the Caribbean coral disease white plague type II (WPII).We then used the probe to test field samples of diseased coral tissue for the presence of this pathogen.Probe design was based on a unique subset (25 nucleotides)of the complete16S rRNA gene sequence derived from a pure culture of the pathogen.The pathogen-specific probe was labeled with the fluorochrome GreenStar*FITC (fluorescein isothiocyanate,GeneDetect Ltd,New Zealand).As a control, we used the universal eubacterial probe EUB 338,labeled with a different fluorochrome (TRITC,tetra-methyl-rhodamine isothiocyanate).Both probes were applied to laboratory samples of pure cultures of bacteria, and field samples collected from the surface of the disease line of corals exhibiting signs of white plague (types I and II),healthy controls,and corals with an uncharacterized disease ("patchy necrosis ").All samples were analyzed using fluorescence in situ hybridization (FISH).We have determined that the probe is specific to our laboratory culture of the coral pathogen,and does not react with other bacterial species (the eubacterial probe does).The WPII pathogen was detected in association with diseased coral samples collected from coral colonies on reefs of the Bahamas (n=9 samples)exhibiting signs of both WPI and WPII.Diseased (and healthy)tissue samples (n=4)from corals exhibiting signs of "patchy necrosis "were also assayed.In this case the results were negative, indicating that the same pathogen is not involved in the two diseases.Incorporation and use of pathogen-specific probes can significantly expand our knowledge of the etiology of coral diseases.
Key words:White plague,coral pathogen,Aurantimonas coralicida,coral diseases,pathogen specific molecular probe.
It is widely believed that coral diseases are significantly contributing to the current,world-wide degradation of coral reefs (Rosenberg and Loya 2004).Since coral diseases were first noted approximately three decades ago,29 individual coral diseases have been proposed (Sutherland et al.2004,Weil 2004).Of these, only five have been characterized to the point that there are known pathogens (Rosenberg and Loya 2004).We are just beginning to investigate how coral pathogens are transmitted between host colonies on the reef,to identify pathogen reservoirs,and to investigate whether or not known pathogens are associated with uncharacterized coral diseases.
One of the most promising approaches to investigating coral pathogens is via the use of pathogen specific molecular probes,a technique that has proven successful in the identification of microorganisms in the environment (Amann 1995).We report here the use of this approach to study a known pathogen of one coral disease,white plague,and to answer some of the current questions concerning the etiology of this disease.
The coral disease white plague,also called plague,was first discovered on reefs of the northern Florida Keys in 1977 (Dustan 1977). It is one of the first three coral diseases reported,along with black band disease (Antonius 1973)and white band disease (Gladfelter et al. 1977).Since its first documented emergence in the 1970s,white plague has reappeared in a much more virulent form on Florida s reefs (Richardson et al.1998a,b,2001)and is now found throughout the Wider Caribbean (Sutherland et al.2004,Weil 2004).
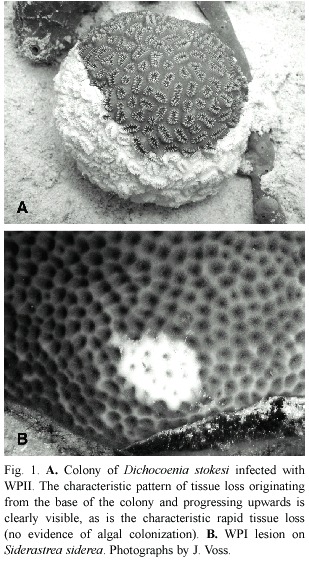
Two recognized forms of plague are now distinguished as white plague (or plague)types I and II.While both present the same clinical signs –a sharp demarcation between apparently healthy coral tissue and freshly exposed coral skeleton (Fig.1)–they differ in the overall patterns and rates of coral tissue destruction,as well as numbers of coral species that are known hosts.White plague type I (WPI)was documented to affect six species of scleractinian corals in the first quantitative study of this coral disease (Dustan 1977).The disease was manifested as lesions occurring on the sides of affected colonies,with an associated rate of progressive tissue destruction of up to 3.1 mm/ day that led to death of entire coral colonies over a period of four months.The most susceptible coral species was Mycetophyllia ferox in the 1970s,while in the 1980s it appeared that Montastraea annularis was most susceptible (Dustan and Halas 1987).While no pathogen was isolated,the disease was transmittable by inoculating healthy colonies with material collected by syringe from infected colonies. Microscopic observation revealed both rod-shaped and flexibacteria associated with the disease line (Dustan 1977).
The second form of the disease,WPII, appeared on the same reefs of the northern Florida Keys in 1995.In contrast to WPI,this form of the disease affected 17 species of scleractinian corals plus the hydrocoral Millipora (Richardson et al.1998b).These included three of the six species reported as affected by Dustan in 1977.In the 1995 outbreak,the most susceptible species was Dichocoenia stokesi. This species was included in Dustan s study on WPI,however 100%of the D.stokesi colonies he examined (n=102)were reported as healthy (Dustan 1977).In contrast,the overall prevalence of WPII within the population of D.stokesi calculated from the 1995 survey (n= 1196 colonies;reef area surveyed=8478 m2 ) was 20.1%,with a range of 0 to 33%infected colonies per site surveyed.The 1995 WPII epizootic resulted in 38%mortality of D.stokesi within populations repeatedly surveyed during a ten-week survey period,which constituted one half of the time period of the epizootic (Richardson et al.1998b).
In addition to differences in coral species affected,WPII exhibited a much faster rate of tissue destruction of up to 2 cm/day (Richardson et al.1998a).Death of individual colonies occurred in two to three days as opposed to months for WPI.Primarily small (<20 cm diameter)colonies of all species were affected,for unknown reasons.WPII also Fig.1.A.Colony of Dichocoenia stokesi infected with WPII.The characteristic pattern of tissue loss originating from the base of the colony and progressing upwards is clearly visible,as is the characteristic rapid tissue loss (no evidence of algal colonization).B.WPI lesion on Siderastrea siderea .Photographs by J.Voss. exhibited a very specific and unique pattern of tissue destruction in that virtually every colony observed to be infected (of which there were many thousands)exhibited tissue loss that started at the base of individual colonies and progressed upwards (Fig.1A).Since the original characterization of this disease in 1995 it has repeatedly occurred as an epizootic in regional areas of reefs of Florida (Richardson et al.1998b)and throughout the Caribbean (Green and Bruckner 2000,Weil et al.2002, Croquer et al.2003,Borger 2003,Miller et al. 2003).It is now reported to affect 40 species of Caribbean corals (Sutherland et al.2004).
Of the two plague types,a pathogen has been isolated from only one,WPII.The pathogen,obtained and isolated from a WPII diseased colony of D.stokesi during the 1995 outbreak, was originally thought to be a possible new species of the bacterial genus Sphingomonas (Richardson et al.1998b).After complete sequencing of the 16S rRNA gene was carried out and additional taxonomically relevant data obtained,we now know that it is a novel genus and species within the order Rhizobiales of the class Alphaproteobacteria (Denner et al.2003). It has been given the name Aurantimonas coralicida Denner et al.,2003,and is a gram negative,polarly flagellated,obligate aerobe.When plated onto marine agar,it forms translucent, compact,shiny colonies that,after two days at room temperature,become pigmented and appear golden yellow.Hence the Latin name of "golden-colored coral killer ".
One question of interest is whether or not A.coralicida is also the pathogen of WPI.It is possible that these two plague etiologies are caused by different strains of the pathogen with different levels of virulence.It is also possible that the two etiologies are caused by entirely different pathogens that infect corals and cause the same disease signs.These hypotheses have been applied to other emerging coral and marine diseases (Harvell et al.1999,Ritchie et al.2001,Bythell et al.2004)yet little research has been carried out to address these issues.In particular,it has been proposed that the same pathogen may very well be involved in the various "white "diseases of corals for which no pathogen has been found.These diseases include,white band disease type I patchy necrosis,"plague-like "disease,and shut-down reaction (Bythell et al.2004).
We report here the development of a species-specific molecular probe for Aurantimonas coralicida .We also present laboratory probe validation results,as well as the first results of use of the probe to test field samples collected from corals with signs of WPI,WPII,and infected corals from an uncharacterized disease outbreak tentatively identified as patchy necrosis.
Materials and methods
Probe design and construction
The A.coralicida specific molecular probe (designated WP)was designed by GeneDetect* based on a unique partial sequence of 25 nucleotides (631-655)located within the A.coralicida 16S rRNA gene (obtained from GenBank accession number AY065627).The (antisense)probe sequence is ACACCAGGTCACTCGGCGGAAGCGG. The probe was designed and constructed by GeneDetect Ltd,New Zealand,and was labeled with the fluorochrome GreenStar* FITC (fluorescein isothiocyanate).The FITC fluorochrome has an excitation at 488 nm (argon laser)and emission at 530 nm.As a control we used a universal eubacterial probe (EUB 338),also constructed by GeneDetect,with the fluorochrome TRITC (tetra-methy-rhodamine isothiocyanate).The TRITC fluorochrome has different excitation (530 nm)and emission (590-620 nm) wavelengths than the WP probe.
Pure culture probe validation
Both the A.coralicida specific probe WP and EUB 338 were validated in the laboratory using the pure culture of A.coralicida (maintained on Difco marine agar slants)and a pure culture of Escherichia coli obtained from ATCC (maintained on Difco nutrient agar slants).
Field sample collection
Samples consisted of surface mucopolysacharide from both healthy and diseased corals, plus lysing coral tissue from diseased corals, collected using sterile 10 ml syringes while SCUBA diving.Samples (n=9)were collected from corals exhibiting signs of either WPI or WPII and included three coral species, Dichocoenia stokesi ,Montastraea annularis, and Siderastraea siderea ,at four sites (Goby Spot,Horseshoe Reef,and North and South Perry Reefs)near Lee Stocking Island,Exuma Chain,Bahamas.Healthy samples were collected from the same (white plague diseased) coral colonies,but from apparently healthy tissue at least 5 cm from the disease line. After sample collection (upon return to the boat),syringes were stored on ice until arrival at the laboratory.At this point,samples were placed in an upright position in ice for 5 min, and mucous/tissue was allowed to settle to the end of the syringe,after which 1.5 ml of the sample containing the mucous/tissue was injected into 1.5 ml sterile cryovials and stored at –40 °C.Samples were hand-carried to Florida International University (FIU)on ice and stored at –20 °C.At no point in time did samples thaw prior to analysis.Of the nine field samples assayed,two were from WPI-diseased tissue,four from WPII-diseased tissue, two from the surface mucopolysaccharide of healthy areas of diseased corals samples,and one from the surface of coral skeleton exposed by WP (Table 1).
Samples were also collected from corals affected by an epizootic that occurred on reefs of Florida in 2003.The disease was tentatively suggested to be patchy necrosis,one of the currently uncharacterized coral diseases,based on disease signs.For our study,samples of lysing tissue from the disease line (along with healthy controls of surface mucopolysaccharide)were collected from Acropora cervicornis colonies at Elkhorn Reef in Biscayne Bay,South Florida.Sterile 10 ml syringes were used,but in this case the entire syringes were immersed in ice immediately upon return to the boat,and maintained (still in the syringes)at –20 °C until analysis at FIU.
Sample preparation
For probe analysis of pure cultures,cells were grown overnight in Marine broth (A.coralicida )or nutrient broth (E.coli )at 32 °C.For probe analysis of field samples,frozen field samples were first thawed.Subsamples (field or laboratory cultures)were first suspended in 0.75 to 1.0 ml PBS buffer (Sigma Dulbecco s Phosphate Buffered Saline)in Eppendorf tubes, and centrifuged at 12 000 rpm for 5 min.The supernatant was removed to leave approximately 100 µl of sample,to which was added 300 µl of 4%paraformaldehyde.Cells were then held at 4 °C for 1.5 hr.Fixed cells were centrifuged at 12 000 rpm for 5 min,the supernatant removed,resuspended in 1 ml PBS,again centrifuged (12 000 rpm for 5 min),and supernatant removed.If a pellet was present,cells were resuspended in 500 µl PBS.If no pellet was present,the overlying liquid was carefully removed by pipette to leave approximately 350 µl of sample in the bottom of the tube.Then, 500 µl of ice-cold 95%ethanol (EtOH)was added,and the cell suspension mixed by pulsing briefly using a vortex mixer.The fixed, suspended cells were spotted (7.5 µl)onto clean microscope slides (previously cleaned with 95%EtOH)and allowed to air dry.Slides were then either immediately analyzed using the probe,or stored overnight at -20 °C.
Probe application and hybridization
Before applying the hybridization buffer,slides were dehydrated in an EtOH series (50%,80%,95%,3 min at each concentration). Eight µl of hybridization buffer (720 µl of 5M NaCl;80 µl of 1M Tris-HCl;1.6 ml formamide;1596 ml autoclaved ddI H2O;and 4 µl 10%SDS)were added to each dried sample spot.Both probes (WP and EUB 338,0.5 µl of each)were added to each sample preparation on the slides and mixed gently with a pipette tip being careful not to disrupt the cell smears. Slides were next placed horizontally in an aluminum foil box (sample face up)on a bed of Kimwipes soaked with extra hybridization buffer,and placed in a hybridization chamber (46 °C)for 1.5-2 hr.After hybridization,slides were washed immediately with 1-2 ml of 55 °C wash buffer (460 µl 5M NaCl and 1 ml 1M Tris-HCl,brought up to 50 ml with autoclaved ddI water with a final addition of 50 µl 10% SDS).Slides,immersed in wash buffer,were held at 55 °C in a temperature-controlled incubator for 15 min.Slides were rinsed briefly in ice-cold dH 2 O,dried by exposure to a controlled airflow,and stored in darkness at 4 ºC until viewed under the microscope.
Fluorescence in situ hybridization (FISH)
Slides were assayed,and hybridization of the probe detected,using a Leica fluorescence microscope equipped with filters for the FITC and TRITC wavelengths,interfaced with computer equipped with an image processing system.Before microscopic analysis,a drop of Citifluor AF1 (Citifluor Ltd.,London)was placed onto the sample (before the cover slip was added)to retard photobleaching of the fluorochromes.Results were recorded by saving images.All results were at a final magnification of 1000x.
Underwater photography
Underwater photographs were taken using an Olympic C-4000 digital camera in an Ikelite housing.
Results
Laboratory validation
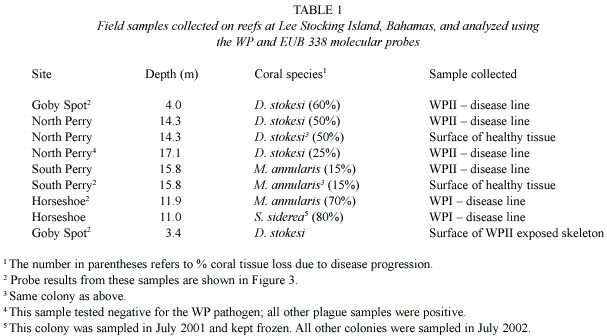
Fig.2 documents the specificity of the molecular probes with pure cultures in the laboratory.In each preparation,both the eubacterial probe (EUB 338)and the A.coralicida probe (WP)were applied to the same preparation on slides.Fig.2A shows a sample of A. coralicida using excitation wavelengths and filters for detection of the WP probe.The result is positive.Fig.2B is the same preparation but using excitation wavelengths and filters for detection of the EUB 338 probe.Again the result is positive,as would be expected.Fig. 2C shows probe analysis of E.coli .EUB 338 was positive (depicted in Fig.2C)while the WP probe did not hybridize (not shown as there was no detectable fluorescence).
White plague field samples
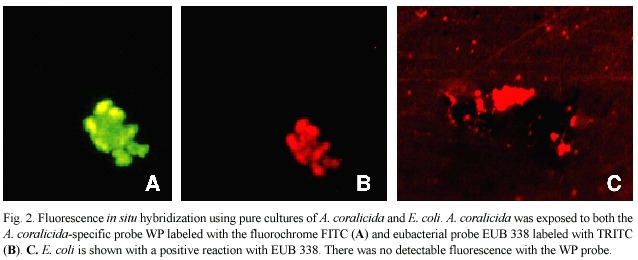
Table 1 summarizes the field samples collected on reefs of Lee Stocking Island, Bahamas,that were analyzed using both molecular probes.Fig.3 shows some of the results of the molecular probe analysis (samples are indicated in Table 1),again using both the WP and EUB 338 probes.Fig.3 includes results of field samples collected from diseased and healthy tissues as well as exposed coral skeleton. Fig.3A,B are from lysed tissue/surface mucopolysaccharide samples of the disease line of a coral colony (Dichocoenia stokesi ) with signs of WPII.Both probe results are positive.Fig.3C,D are from lysed tissue/surface mucopolysaccharide samples of the disease line of a coral colony (Montastraea annularis ) with signs of WPI.Again both probe results are positive.Fig.3 E,F,G show results of the probe analysis of healthy surface tissue/muco-polysaccharide from a colony of M.annularis with WPII (sample collected away from the disease line).In these images,3E is excited by the wavelengths specific to the WP probe and 3F by wavelengths specific to the EUB 338 probe.Fig.3G had no transmittance filter, and the only available light was leakage from the halogen light source.Thus 3G reveals the general fluorescence of coral tissue.Fig.3H is a surface sample of coral skeleton (again D. stokesi )freshly exposed by WPII.The sample tested positive (only WP probe shown).
We obtained one negative result from the nine field samples assayed –one sample collected from a WPII disease line (indicated in Table 1)was not positive for the WP probe (but was for EUB 338).
Patchy necrosis field samples
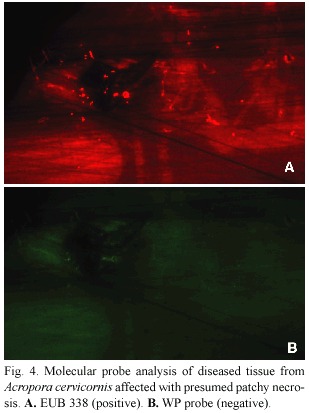
Neither diseased nor healthy samples from A. cervicornis infected with presumed patchy necrosis tested positive for the WP probe (Fig.4).
Discussion
The molecular probe we used in this study was constructed from a culture of A.coralicida originally isolated from a coral infected with WPII (Richardson et al.1998b).This form of plague has an etiology characterized by more rapid tissue lysis than that of WPI. Our results indicate that the same pathogen is involved in both plague etiologies.The difference in rate of tissue lysis may be due to expression of an increased level of a virulence factor (or factors)by the pathogen in WPII. These findings thus serve as a basis for future comparative virulence studies of pathogen activity in WPI vs.WPII.
We also obtained positive probe results in a sample collected from a coral skeleton that was recently exposed by white plague, revealing at least one potential reservoir for A. coralicida in addition to infected coral tissue. Further investigation of possible reservoirs of this pathogen can now be conducted to more clearly define the epizootiology of WP.
The probe was also used to assess samples collected from Florida Reefs during an outbreak of a coral disease that occurred in July 2003. This uncharacterized disease was tentatively identified as "patchy necrosis ".It was present on colonies of both Acropora palmata and A. cervicornis .Patchy necrosis is reported in the literature (Weil 2004)as affecting a single host species (A.palmata ),therefore there is some question as to the identification assigned to this disease. However, other coral diseases (including WP)have exhibited rapid expansions of host range,which could potentially be occurring in this case.A pathogen has not been found for patchy necrosis (Rodriguez-Martinez et al. 2001).Our assessment of samples from this outbreak was carried out to test the hypothesis that there may be a common pathogen among the uncharacterized "white "diseases,which include patchy necrosis and "plague-like "diseases (Bythell et al.2004).We were able to show that A.coralicida is not associated with the "patchy necrosis "outbreak in 2003. The probe would also prove useful in the study of additional,uncharacterized diseases. One such recently described coral disease was characterized by patchy lesions on scleractinian coral colonies (Bythell et al.2002).Although the lesions did not fit the published descriptions of white plague from either Dustan (1977)or Richardson et al.(1998a),it was termed white plague (Bythell et al.2002)based on Dustan s original definition of plague as a potential "suite of diseases "(Dustan 1977).Histopathology of samples from this disease revealed coccoid bacteria of a different shape than the rods formed by A.coralicida .The identity of these bacteria was not assessed genetically (non-cultivative)or via culturing and isolating.Yet another disease investigated by this group, termed "plague-like "(Pantos et al.2003),was examined using molecular (16S rRNA gene sequence)analysis.In this case,results indicated the presence of alphaproteobacteria from the Roseobacter/Rhodobacter group.There was no sequence homology to A.coralicida. In each of the above cases, application of the WP probe to samples of diseased tissue would provide a definitive yes or no answer to the question of whether or not A.coralicida is an associated pathogen.
In conclusion,use of species-specific molecular probes has provided information about the identity of coral pathogens associated with three coral diseases:WPI,WPII,and an uncharacterized disease tentatively identified as patchy necrosis.We have been able to demonstrate that corals exhibiting signs of white plague types I and II are both infected by A.coralicida ,the WPII pathogen,and that "patchy necrosis "on A.cervicornis was not. Further application of the WP probe will be valuable for the quantitative study of other unanswered questions concerning white plague epizootiology.For example,we do not yet know the identity of the white plague pathogen reservoir.We do not understand why epizootics of WPII,common now throughout the Caribbean,exhibit a pattern in which an Fig. 4. Molecular probe analysis of diseased tissue from Acropora cervicornis affected with presumed patchy necrosis.A.EUB 338 (positive).B.WP probe (negative).
Acknowledgments
We thank Kalai Mathee for use of the epifluorescence microscope and equipment in her molecular biology laboratory,and Travis Thyberg and Ashley Ford for assistance in field sampling at Lee Stocking Island.The "patchy necrosis "samples were provided within a collaboration coordinated by the Coral Disease and Health Consortium,and we thank Cheryl Woodley,Margaret Miller,Dana Williams, Samantha Ryan,Shawn Polson and Mats Lundqum for this effort.This paper is a result of research funded by the National Oceanic and Atmospheric Administration Coastal Ocean Program under award number NA16OA1443 to the National Coral Reef Institute at Nova Southeastern University (to LR and DM), by a grant from the Caribbean Marine Research Center (Project CMRC-02-PRJV- 01-02A)National Oceanic and Atmospheric Administration (NOAA)Undersea Research Program (to J.V and L.R),and by an EPA Fellowship to J.V.Views expressed herein are those of the authors and do not necessarily reflect the views of CMRC,NOAA,or any of their sub-agencies.This is contribution number 86 from the Tropical Biology Institute of Florida International University.
Resumen
Uno de los problemas actuales en el campo de la investigación sobre las enfermedades de corales es el de poder seguir a los patógenos en el ambiente natural.Un método prometedor para lograrlo es el uso de sondas moleculares específicas para el patógeno.Sin embargo,esta técnica ha sido poco usada hasta la fecha.Construimos y validamos en el laboratorio una sonda molecular de fluorocromo marcado específicamente para Aurantimonas coralicida ,la bacteria responsable de la enfermedad de corales del Caribe,plaga blanca Tipo II (PBII).Después usamos la sonda con muestras de campo de tejidos de corales enfermos para detectar la presencia del patógeno.La sonda se diseñó usando un sub-grupo único de 25 nucleótidos del gen 16S del rARN derivado de cultivos puros del patógeno.La sonda específica para el patógeno se marcó con fluorocromo GreenStar* FITC (fluorescein isotiocianato,GeneDetect Ltd,New Zealand).Como testigo usamos la sonda universal de eubacterias EUB 338,marcada con un fluorocromo diferente (TRITC,tetra-metil-rodamina isotiocianato).Ambas sondas fueron usadas con muestras de laboratorio de cultivos puros de bacterias,y muestras de campo recolectadas de la superficie de la línea de la enfermedad en corales con signos de plaga blanca (tipos I y II),de corales sanos y de corales con enfermedades no características (parches de tejido necrótico).Todas las muestras fueron analizadas usando hibridización fluorescente in situ .Determinamos que la sonda es específica para el patógeno cultivado en nuestro laboratorio y no reacciona con otras especies de bacterias (la sonda para eubacterias sí).El patógeno de PBII fue detectado en muestras de corales enfermos (exhibían signos de PBI y PBII)recolectadas en arrecifes de Bahamas (n=9 muestras).Muestras de tejidos enfermos (y sanos)(n=4)de corales con parches necróticos fueron probados.En este caso los resultados fueron negativos,indicando que el mismo patógeno no es responsable de las dos enfermedades.El desarrollo y uso de sondas específicas para cierto patógeno puede ampliar significativamente nuestro conocimiento de la etiología de enfermedades de corales.
Palabras clave:Plaga blanca,patología de corales, Aurantimonas coralicida,enfermedades de coral,sonda molecular para patógeno específico.
References
Amann,R.I.1995.In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes,p.1-15.In A.D.L.Akkerman, J.D.van Elsas &F.J.de Bruijn (eds.).Molecular Microbial Ecology Manual.Kluwer Academic Publishers,Dordrecht,The Netherlands. [ Links ]
Antonius,A.1973.New observations on coral destruction in reefs.10th Mtg.Assoc.Isl.Mar.Lab.Carib. 10:3. [ Links ]
Borger,J.2003.Three scleractinian coral diseases in Dominica,West Indies:Distribution,infection patterns,and contribution to coral tissue mortality.Rev. Biol.Trop.5 (Supl.4):25-38. [ Links ]
Bythell,J.,M.Barer,R.Cooney,J.Guest,A.O Donnell, O.Pantos &M.Le Tissier.2002.Histopathological methods for the investigation of microbial communities associated with disease lesions in reef corals. Lett. Appl.Microbiol.34:111-116. [ Links ]
Bythell,J.,O.Pantos &L.Richardson.2004.White plague, white band,and other "white "diseases,p.351-365. In E.Rosenberg &Y.Loya (eds.).Coral Health and Disease.Springer-Verlag,Berlin. [ Links ]
Croquer,A.,S.M.Pauls &A.L.Zubillaga.2003.White plague disease outbreak in a coral reef at Los Roques National Park,Venezuela.Rev.Biol.Trop.5 (Supl. 4):39-45. [ Links ]
Denner,E.B.M.,G.W.Smith,H-J Busse,P.Schumann,T. Narzt,S.Polson,W.Lubitz &L.L.Richardson.2003. Aurantimonas coralicida gen.nov.,sp.nov.,the causative agent of white plague type II on Caribbean scleractinian corals. Intl. J. Syst. Evol.Microbiol.53: 1115-1122. [ Links ]
Dustan,P.1977.Vitality of reef coral populations off Key Largo,Florida:Recruitment and mortality. Env.Geol. 2:51-58. [ Links ]
Dustan,P.&J.Halas.1987.Changes in the reef-coral community of Carysfort Reef,Key Largo,Florida: 1974-1982.Coral Reefs 6:91-106. [ Links ]
Gladfelter,W.B.,E.H.Gladfelter,R.K.Monahan, J.C.Ogden,&R.F.Dill.1977.Environmental studies of Buck Island Reef National Monument,St.Croix US Virgin Islands.Spec.Rep.Nat.Park Serv.US Dept Int.173 p. [ Links ]
Green,E.P.&A.W.Bruckner.2000.The significance of coral disease epizootiology for coral reef conservation. Biol. Cons. 96:347-361. [ Links ]
Harvell,C.D.,K.Kim,J.M.Burkholder,R.R.Colwell, P.R.Epstein,D.J.Grimes,E.E.Hofmann,E.K.Lipp, A.D.M.E. Osterhaus, R.M.Overstreet,J.W.Porter, G.W.Smith &G.R.Vasta.1999.Emerging marine diseases -climate links and anthropogenic factors. Science 285:1505-1510. [ Links ]
Miller,J.,C.Rogers &R.Waara.2003.Monitoring the coral disease,white plague type II,on coral reefs in St.John,U.S.Virgin Islands.Rev.Biol.Trop.5 (Supl.4):47-55. [ Links ]
Pantos,O.,R.Cooney,M.Le Tissier,M.Barer,A. O Donnell &J.Bythell.2003.The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastraea annularis .Env.Microbiol.5:370-382. [ Links ]
Richardson,L.L.,W.M.Goldberg,R.G.Carlton &J.C. Halas.1998a.Coral disease outbreak in the Florida Keys:Plague type II.Rev.Biol.Trop.46 (Supl.5): 187-198. [ Links ]
Richardson,L.L.,W.M.Goldberg,K.G.Kuta,R.B. Aronson,G.W.Smith,K.B.Ritchie,J.C.Halas,J.S. Feingold &S.L.Miller.1998b.Florida s mystery coral killer explained.Nature 392:557-558. [ Links ]
Richardson,L.L.,G.W.Smith,K.B.Ritchie &R.G. Carlton.2001.Integrating microbiological,microsensor,physiologic and molecular techniques in the study of coral disease pathogenesis.Hydrobiol.460: 71-89. [ Links ]
Ritchie,K.B.,S.W.Polson &G.W.Smith.2001.Microbial disease causation in marine invertebrates:Problems, practices,and future prospects.Hydrobiol.460: 131-139. [ Links ]
Rodriguez-Martinez,R.E.,A.T.Banaszak &E.Jordan- Dahlgren.2001.Necrotic patches affect Acropora palmata (Scleractinia:acroporidae)in the Mexican Caribbean.Dis.Aquat.Org.47:229-234. [ Links ]
Rosenberg,E.&Y.Loya.2004.Coral Health and Disease. Springer-Verlag,Berlin.488 p. [ Links ]
Sutherland,K.P.,J.W.Porter &C.Torres.2004.Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar.Ecol.Prog.Ser.266:273-302. [ Links ]
Weil,E.2004.Coral reef diseases in the wider Caribbean, p.35-68.In E.Rosenberg &Y.Loya (eds.).Coral Health and Disease.Springer-Verlag,Berlin. [ Links ]
Weil,E.,I.Urreiztieta &J.Garzon-Ferreira.2002. Geographic variability in the incidence of coral and octocoral diseases in the wider Caribbean.Proc.9th Int.Coral Reef Symp.2:1231-1238. [ Links ]














