Revista de Biología Tropical
versión On-line ISSN 0034-7744versión impresa ISSN 0034-7744
Rev. biol. trop vol.52 no.3 San José sep. 2004
A phylogeny of howler monkeys (Cebidae: Alouatta) based on mitochondrial, chromosomal and morphological data
Federico Villalobos1,2 , Alejandro A. Valerio1 & Axel P. Retana1
1 Instituto Centroamericano para la Investigación en Biología y Conservación (ICIBC). P.O. Box 2398-2050 San Pedro de Montes de Oca, San José, Costa Rica.
Abstract
The current taxonomic status of the species and subspecies belonging to the genus Alouatta is addressed by combined phylogenetic analysis using morphological, kariotipyc and molecular data (mitochondrial genes cytocrome oxidase II and cytochrome B). Our result demonstrated that Alouatta palliata is the most basal taxon for the genus in concordance with previous studies, as well as showing the validity of the taxon Alouatta sara as a species. Also our analysis shows that the sex chromosome has evolved from a XY/XX system to a X1X2Y1Y2/X1X1X2X2 system within the genus, as well as an increase in the size and complexity of the hioideal bone. Rev. Biol. Trop. 52(3): 665-677. Epub 2004 Dic 15.
Key words: Alouatta, phylogeny, Platyrrhini, taxonomy, hyoid bone, sex chromosome system.
Palabras clave: Alouatta, filogenia, Platyrrhini, taxonomía, hueso hioideo, cromosomas sexuales.
Received 29-IV-2004. Corrected 24-VIII-2004. Accepted 24-VIII-2004.
The evolutionary relationships of the infraorder Platyrrhini (Primates) genera have been studied using morphological (Rosenberg 1981, 1984, Ford 1986, Zingeser 1973, Ford and Davis 1992), karyotypic and molecular data (Schneider et al. 1993, Horovitz et al. 1998, Dutrillaux et al. 1986). Some incongruence between morphological and molecular data has emerged in some groups, such as the subfamily Atelinae (Lagothrix É. Geoffroy 1812, Brachyteles Spix 1823, Ateles É. Geoffroy 1806, and Alouatta Lacépède 1799). The clade Atelinae is considered to be a monophyletic group based on the large body size, a prehensile tail with a hairless ventral area for grasping, the frequent use of suspensory positional behaviors, and other characters (Horowitz et al. 1998, Rosenberg 1989).
The phylogenetic relationships among the Atelinae genera have not been explored until recently (Horovitz et al. 1998). According to Rosenberg (1981, 1984), Ford (1986), Zingeser (1973) and Ford and Davis (1992), reported that the genus Alouatta is the sister clade to a clade compose of Lagothrix, Brachyteles and Ateles based on morphological data (Brachyteles and Ateles are the sister clade of Lagothrix). In contrast, Kay (1990) found the clade Lagothrix + Ateles as the sister clade of Alouatta + Brachyteles based on craneo- dental characters.
Dutrillaux et al. (1986) inferring chromosome rearrangements, reported that Brachyteles, Lagothrix and Ateles form a separate and distinct evolutionary branch from which Lagothrix is more closely related to Ateles than Brachyteles. On the other hand, Schneider et al. (1993, 1996), using DNA sequences for the a-globin and IRBP intron-1, found that Alouatta is the sister taxon to the clade ((Brachyteles+Lagothrix) Ateles). These relationships were tested by Horovitz et al. (1998) in a combined analysis of morphological and molecular data (nuclear and mitochondrial sequences), with the resulting topology identical to the one already obtained by Schneider et al. (1996) for the Atelinae.
The Atelinae subfamily has also been studied at the species level. Froehlich et al. (1991) used a multivariate approach to examine Ateles cranial and dental morphometric patterns and concluded that this genus may be composed of three species. A more recent study using cytogenetical data (Madeiros et al. 1997), however concluded that Ateles fusciceps maybe reproductive isolated and therefore considered as a separated species. The genus Alouatta is currently considered to be composed of 9 species: Alouatta belzebul (Linnaeus 1766), A. seniculus (Linnaeus 1766), A. caraya (Humboldt 1812), A. fusca (É. Geoffroy 1812), A. palliata (Gray 1849), A. pigra (Lawrence 1933), A. sara (Elliot 1910), A. coibensis (Thomas 1902), and A. arctoidea (Rylands 1995). Of those, A. sara and A. arctoidea were subspecies of A. seniculus but now considered as full species by Minezawa et al. (1985) and Stanyon et al. (1995); while A. coibensis was also proposed as a separate species from the formerly known A. palliata panamensis (Froehlich and Froehlich 1987).
In addition, the taxonomic status of A. seniculus macconelli and A. s. straminea is in debate. According with Hill (1962), A. s. macconelli and A. s. straminea were considered as part of the Alouatta seniculus 9 subspecies group. Bonvicino et al. (1995), however, proposed that A. s. seniculus, A. s. stramineus and A. s. macconelli should be considered as separate species basing their results in the analysis of quantitative cranial traits. In contrast, studies done by Figueiredo (1998) using mitochondrial DNA and Sampaio et al. (1996) using biochemical data (20 protein loci) showed that A. s. macconelli and A. s. stramineus are related closely enough to be considered a single subspecies. In addition, Lima and Seuanez (1991) found the same closeness between these two subspecies, but in this case the authors did not support the fusion of these subspecies.
The first systematic revision for the genus Alouatta was conducted by Hershkovitz (1949) using morphological characters. Hershkovitz (1949) proposed its division in three subgroups based on the hyoideal bone morphology, and their composition is as follows: the seniculus group (A. seniculus, A. belzebul, A. fusca), the palliata group (A. palliata and A. pigra) and finally the caraya group (A. caraya). More recently, Sampaio et al. (1996) studied the taxonomy of the Seniculus group containing the species A. seniculus and A. belzebul, based on biochemical and chromosome data. Oliveira (1996) using cytogenetical data proposed the placement of A. belzebul nigerrima together with A. seniculus. Also Gregorin (1996) proposed a phylogenetic hypothesis for the Alouatta species using cranial and hiodeal bone morphology which resulted in the placement as species of most of the A. seniculus subspecies, some of which (A. seniculus ululata, A. s. discolor) were closely related to A. belzebul. The recent study realized by Bonvicino et al. (2001) using Cytochrome B sequences partially support the phylogenetic relationships found previously: A. palliata as the basal taxon to all Alouatta species.
The goal of the present paper is to estimate the phylogenetic relationships among Alouatta species using karyotypic, morphological and DNA data on a simultaneous analysis.
Material and methods
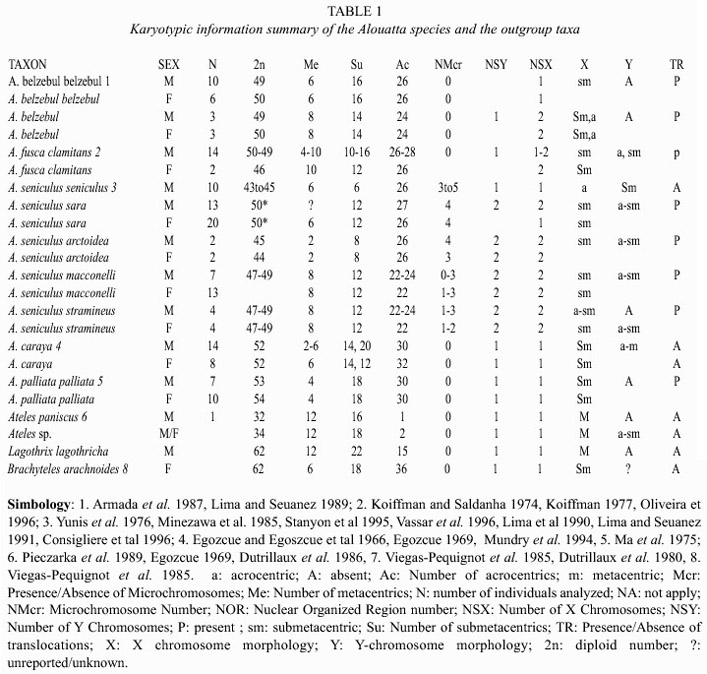
Data collection: The karyotypic data was compiled from existing literature (Table 1). Testing for chromosomal homologies was not possible because the low-resolution techniques used in most of the studies (chromosome homologies only reported by Mudry et al. 1994). Characteristics of the hyoideal bone were evaluated and taken from the illustrations of Hershkovitz (1949) and illustrations by Gregorin (1996); some of the Gregorin matrix characters used in this analysis as well as the hyoideal volumetric data previously reported by Crockett and Eisenberg (1986).
DNA sequences from mitochondrial cytochrome B gene (CYTB) and cytochrome oxidase II (COII) were obtained from genebank and other sources (Table 2). DNA sequence alignments were performed using CLUSTAL W, ver. 1.75. (Thompson et al. 1994) and each gene was independently aligned and then concatenated. Because the genes are from independent studies, the sequences were not from the same localities, therefore the criterion used to concatenate was geographical proximity. The karyotypic and morphological data were added a posteriori to each taxon therefore all the sequences for a species had the same information. For Alouatta pigra and A. coibensis, no available data have been reported from the revised literature.
Phylogenetic analysis and sequence alignment: The phylogenetic analyses for the simultaneous combined matrix were performed using PIWEE, ver. 2.83. (Goloboff 1993). The search strategy consisted in 10000 TBR replicates (mult*), keeping 2 trees by replicate. All the uninformative characters were removed before the analysis. The Bremer decay index was calculated using a sample of 15000 suboptimal trees up to 15% less fit. The preformed homogeneity tests (I.L.D.L test by Farris et al. 1995) were also performed using Winclada ver. 1.00.08 with 1000 replicas.
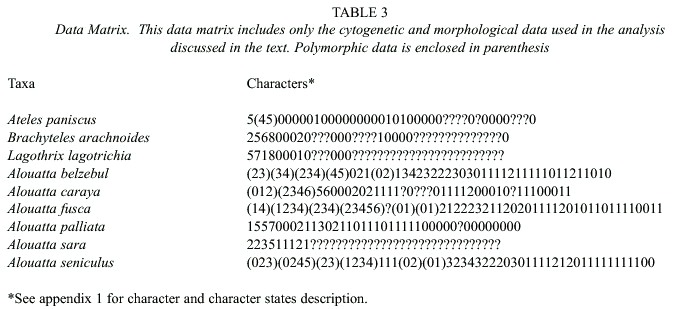
According to White (1978) the karyotypic information for some taxa shows some discernible directional tendencies but all the characters were stated as unordered because in most cases the available information lacked the resolution needed to infer direction for the chromosomal changes. The genera Lagothrix, Brachyteles, and Ateles were used as the outgroup, and their relationships to Alouatta followed the phylogenetic hypotheses proposed by Horowitz et al. (1998) and Schneider et al. (1993). Some morphological characters were treated as additive (Table 3 and Appendix for karyiotipic and morphological part of the matrix).
We choose the use of simultaneous analysis of different data sets because it could improve the phylogenetic signal because of the possible common shared history of different data sets. The homoplasy distribution is likely to be different in each data set because they are subject to different constrains, therefore when different data sets are analyzed simultaneously the signal common to all of them is more likely to overwhelm the homoplasy signal on the data (Kluge 1989).
Results
The molecular partition was found to be incompatible with the morphological partition according to the I.L.D. test (Farris et al. 1975) (P value= 0.0102) as well as the cytochrome oxidase II gene with respect to the cytochrome B gene (P value= 0.0102). According to Cunningham (p.739, 1997), however, when the P value of the IDL test is greater than 0.01 the combination of the "incompatible" data sets improved, or in the worst case did not affected the accuracy of the phylogenetic reconstruction.
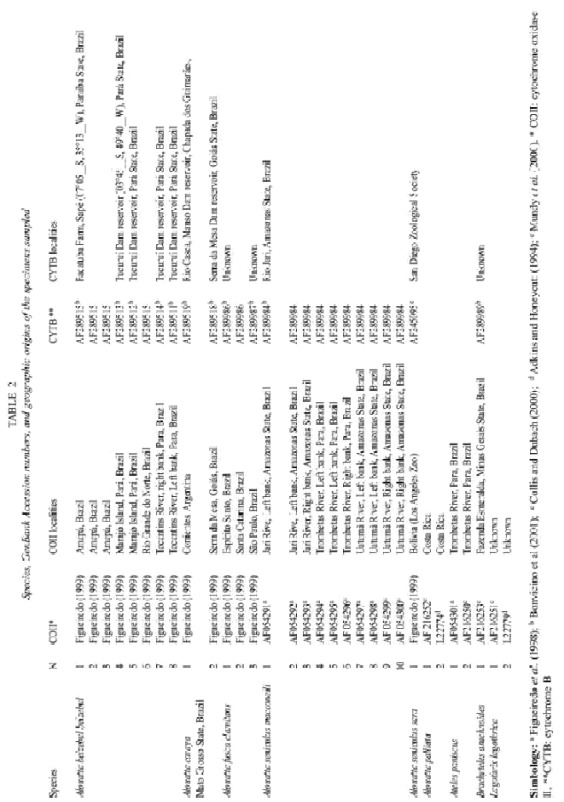
From the 1675 characters used, only 264 were informative. The phylogenetic analysis using the whole data set resulted in 3 equally parsimonious trees from which a strict consensus was calculated (Fig. 1, Length= 463, CI= 0.69; RI= 0.90, RC= 0.621, Fit value= 2315.0). All the obtained trees only differ on the relationships among DNA sequences within the taxon A. seniculus macconelli.
A. palliata is placed as the sister taxon to all other Alouatta species. In addition, A. s. sara is placed as the sister taxon of A. seniculus macconelli, more than 15 fit units would needed to collapse all A. s. macconelli to conform a single clade with A. s. sara (Fig.1). Also, the phylogenetic relationship among A. fusca, A. belzebul and the clade conformed by A. s. sara and A. s. macconelli on the consensus appears to be resolved. 4.5 fit units would be needed to collapse the clade conformed by A. belzebul, A. s. sara and A. s. macconelli (Fig. 1).
Discussion
Taxonomic implications: According to Hershkovitz (1949), the Alouatta genus could be divided in three different groups based on the hyoid bone morphology. These are: the seniculus group (A. seniculus, A. belzebul and A. fusca); the palliata group (A. palliata) (Mittermeier et al. 1988, included A. pigra and A. coibensis); and finally the caraya group (A. caraya). Later, Sampaio et al. (1996), using biochemical and chromosome data, found that A. seniculus and A. belzebul are not the closest species at the seniculus group. Meanwhile, the chromosome data compiled by them supports the notion of a closest relationship between A. belzebul and A. fusca.
Recent independent molecular analysis using the pseudogene a-1-globin (Meireles et al. 1999) and cytochrome B sequences (Bonvicino et al. 2001) produced partially congruent topologies to the one found with the combined data approach. These studies did not included all the species of the genus, therefore A. palliata was never included, not was A. sara. Figueiredo (1998) using the COII gene included this species in her analysis, the trees found could not resolve the relationships among A. belzebul, A. fusca and a polytomy conformed by A. seniculus, A. sara, A. nigerrima and A. caraya. Figueredos (1998) analysis placed A. palliata as the sister taxa to all other species. Morphological analysis based on the hyoid bone was also incongruent with the molecular studies: Gregorin (1999) found A. belzebul and A. seniculus conforming a clade with A. fusca as the sister taxon. Our results support this relationship (Fig. 1).
Our results placed A. palliata as the sister taxon to the remainder taxa agreeing with previously reported topologies (Meireles et al. 1999, Bonvicino et al. 2000). Moreover, A. sara was the sister taxon to A. seniculus macconelli, with a strong support (decay index value of 14.5), confirming its separate species status as previous works (i.e. Minezawa et al.1985, Stanyon et al. 1996). Stanyon et al. (1996) sugested the specific status based on more than 10 chromosomal rearrangements between A. s. sara and A. s. arctoidea.
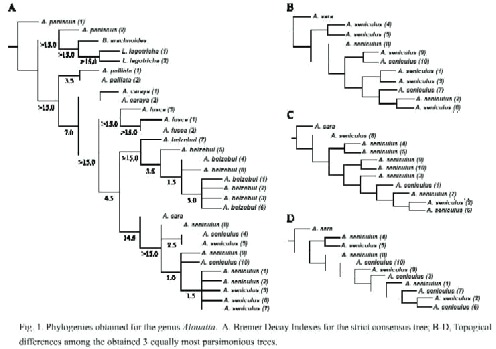
Hyoid evolution: The hyoid bone is a large thin walled subglobular capsule that acts as a resonator and amplifier of Alouattas characteristic loud calls during territorial delimitation and appears to be a very specialized structure (Hershkovitz 1949, Crockett and Eisemberg 1986, Rosenberg 1989). Also, it has been considered a key character for Alouattas species identification (Hershkovitz 1949). There have been few studies so far discussing the evolution of this structure.
In general, our results suggest a tendency towards an increasing complexity for the hyoid bone. There has been a trend towards a more globular hyoid bone, which is observed as an increase in the volume of the bone (with its highest volume exhibited by A. seniculus). Also, the tentorium has undergone a size increase in A. seniculus (Fig 3). The hyiod opening, the cornum and corniculum has been reduced most probably as a consequence of the increasing bulla development (i.e. the hioyd lateral shape changed from a "J" shape in A. palliata to a globular structure in A. seniculus).
The mentioned morphological modifications could account for the present acoustic differences among the species calls. In A. seniculus, the entering of air acoustically activated air by the glottis into the hyoid bulla and lateral air sacs amplify some frequencies, and increase the amplitude of the calls (Schon-Ybarra 1986). The A. seniculus barks range from 350 to 3500 Hz, with a spectral energy concentration on 350-1100 Hz (Schon-Ybarra 1986) while in A. caraya, which lacks of a complex development in the hyoid bone, produce lower call frequencies. The strongest frequencies in barks and long calls of A. caraya are in the range of 310-328 Hz (Thorington et al. 1984). Also, A. palliata, which is the species with the least complex hyoid bone, produces barks and roars at lower frequencies than A. caraya and A. seniculus (Eisenberg 1976, Thorington et al. 1984, Schön-Ybarra 1986).
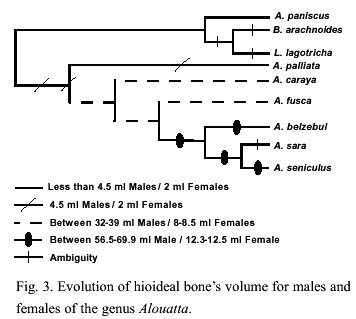
Karyotypic evolution: Several tendencies were found for the Alouatta species like the reduction in the diploid number, which is seen as a further gain for A. caraya and A. palliata (Fig. 2). These tendencies have also been observed in Ateles species (2n= 34-33) in comparison with the genera Lagothrix and Brachyteles who have a diploid number of 62. Pieczarka et al. (1989) reported for Ateles paniscus paniscus a similar situation as a result of the fusion between the chromosomes 4 and 12. Fusion of chromosomes was also found in A. belzebul belzebul, A. b. hybridus, and A. geoffroyi (Garcia et al. 1975, Benirshke 1975, Kunkel et al. 1980). Nevertheless, it is important to mention that the diploid number for the species is the result of a population sampling process, making this character extremely variable (Ma et al. 1975).
The sex chromosome system in the genus Alouatta is highly diverse in comparison with other Neotropical primates. A. palliata has been described to have an XY system for males and XX for females (Ma et al. 1975) from a population studied in Barra Colorado Island (Panama). A. caraya has also been reported to have this same sex chromosome system (Oliveira 1996). A. fusca, however, shows at least 4 different karyomorphic groups: 2n=52 XY/XX; 2n= 48 or 50 XY/XX; 2n=49, X1X2Y/50,X1X1X2X2 and 2n=45 X1X2Y/ 46 X1X1X2X2 (Oliveira et al. 1995, 1998). It seems like the origin of these groups in A. fusca appear independently where the de novo sex chromosomes came from different autosomes (Bonvicino et al. 2001) A. belzebul also present a similar sex chromosome system of 50 X1X1X2X2/49X1X2Y (Armada et al. 1987), while A. macconelli, A. sara and S. arctoidea showed a X1X2Y1Y2/X1X1X2X2 system (Consigliere et al. 1996, Stanyon et al. 1995, Lima et al. 1990).
According with our results, the sex chromosome has evolved from a XY/XX system to a X1X2Y1Y2/X1X1X2X2 system. A. fusca is a polymorphic species with regard to the sex chromosome system, and due to its phylogenetic position in our results it was expected to present a XY/XX system. Due to the polymorphic status of this species it is not possible to asset unambiguously a character state at the node shared with the clade composed by A. belzebul, A. sara and A. macconelli.
The existence of microchromosomes in some species of the genus Alouatta has been another distinctive feature. These chromosomes have been found in A. sara, A. s. artoidea (Consigliere et al. 1996, Minezawa et al. 1985), A. s. seniculus (Yunis et al. 1976) and A. s. macconelli (Lima et al. 1990, Vassart et al., 1996). For A. palliata (Ma et al. 1975), A. caraya (Egozcue and Egozcue 1966, Mudry et al. 1994, Egozcue 1969) and A. belzebul belzebul (Armada et al. 1987), however the presence of microchromosomes has not been reported. In A. fusca, the absence of microchromosomes has been reported (Koiffman 1982, Koiffman and Saldanha 1974), but the authors mention a numerical variation of karyotype in the number of the smallest pair. Minezawa et al. (1985) pointed out that this variation could be due to the presence of "accessory chromosomes" (microchromosomes). According with the present results, if the absence of microchromosomes in A. fusca is considered, the microchromosomes would have evolve once in the genus for the taxa A. sara and A. seniculus. Conversely, if the numerical variation on the karyotype of A. fusca is considered as proof of the microchromosome presence then the microchromosomes would have acquired independently by A. fusca and the clade A. sara and A. seniculus.
Acknowledgments
We like to thank the following people who provided support and comments: Cukha, Dione, Santiago, Antonia, S. Cameron, Sascha Pfennig and J.B. Whitfield. This research was supported by the Central American Institute for Biological Research and Conservation grand number CIBRC 0001-A.
Resumen
El estado taxonómico actual de las especies y subespecies del genero Alouatta (Lacépède, 1799) fue estudiado empleando un análisis filogenético combinado de datos morfológicos, cariotipicos y moleculares (genes mitocontridales del Citocromo Oxidasa II y el Citocromo B). Nuestros resultados demuestran que Alouatta palliata (Gray 1949) es la especie mas basal del genero en concordancia con propuestas previas para el grupo, también muestran la valides de Alouatta sara (Elliot 1910) como una especie. Nuestros análisis también muestra que los cromosomas sexuales evolucionaron de un sistema XY/XX a un sistema X1X2Y1Y2/X1X1X2X2 dentro del genero así como también un incremento en el tamaño y complejidad del hueso hioideo.
References
Adkins, R. & R.L. Honeycutt. 1994. Evolution of the primate cytochrome c oxidase subunit II gene. J. Molec. Evol. 38: 215-231. [ Links ]
Armada, J.L.A., C.M.L. Barroso, M.M.C. Lima, J.A.P.C. Muniz & H.N. Seuanez. 1987. Chromosome studies in Alouatta belzebul. Am. J. Primatol. 13: 283-296. [ Links ]
Benirshkle, K. 1975. Biomedical research. In. Research in zoos and acuariums. Ed. by Institute of laboratory animal resources. Nat. Acad. Sci. Washington DC. 3-11. [ Links ]
Bonvicino, C.R., M.E.B. Fernades & H.N. Seuanez. 1995. Morphological analysis of Alouatta seniculus species group Primates. Cebidae. Acomparison with biochemical and karyological data. Hum. Evol. 10: 169-176. [ Links ]
Bonvicino, C.R., B. Lemos & H.N. Seuanez. 2001. Molecular phylogenetics of the howler monkey Alouatta Platyrrhini. a comparison with karyotypic data. Chromosoma 110: 241-246. [ Links ]
Collins, A.C. & J.M. Dubach. 2000. Phylogenetic Relationships of Spider Monkeys Ateles Based on Mitochondrial DNA Variation. Int. J. Primatol. 21: 381-420. [ Links ]
Consiliglieri, S., R. Stanyon, U. Koehler, Agoramoorthy G. & J. Wienberg. 1996. Chromosome painting defines genomic rearrangements between red howler monkey subspecies. Chrom. Res. 4: 264-270. [ Links ]
Crockett, C.M. & J.F. Eisenberg. 1986. Howlers. variation in group sizes and demography. In B.B. Smuts, D.L. Cheney, R.M. Seyfarth R.W. Wrangham & T.T. Struhsaker (eds.). Primate societies. University of Chicago, Chicago USA. [ Links ]
Cunningham, C.W. 1997. Can tree incongruence test predict when data should be combined? Mol. Biol. Evol. 14: 733-740. [ Links ]
Dutrillaux, B. J. Couturier & A.M. Fosse. 1980. The use of high resolution banding in comparative cytogenetics. comparizion between man and Lagothrix lagotricha (Cebidae). Cythogen. Cell. Gent. 27: 45-51. [ Links ]
Dutrillaux, B., J. Couturier & E.Viegas-Pequignot. 1986. Evolucion cromosomique des Platyrhiniens. Mammalia 50: 56-81. [ Links ]
Eisenberg, J. 1976. Comunication mechanisms and social integration in the black spider monkey Ateles fusciceps robustus and related species. Smithson. Contr. Zool 213: 1-108. [ Links ]
Elliot, D.G. 1910. Description of new species of monkeys of the genera Galogo Cebus Alouatta and Cercopithecus. Ann. Mag. Nat. Hist. 8: 77-83. [ Links ]
Egozcue, J. & M.V. Egozcue. 1966. The chromosime complement of the howler monkey Alouatta caraya Humboldt 1812. Cythogenetics 5: 20-27. [ Links ]
Egozcue, J. 1969. Primates. In K. Benirschke (Ed.). Comparative mammalian cytogenetics. Springer, New York USA. [ Links ]
Farris, J.S., Kallersjo M. & C. Bult. 1995. Constructing a Significance Test for Incongruence. Syst. Biol. 44: 572. [ Links ]
Figueiredo, W.B. 1999. Filogenia molecular do genero Alouatta Lacepede (1799) Atelidae. Primates. MSc Dissertation Universidade Federal do Para Para Brazil. [ Links ]
Figueiredo, W.B, N.M. Carvalho-Filho, H. Schneider & I. Sampaio. 1998. Mitochondrial DNA sequences and the taxonomic status of Alouatta seniculus populations in northeastern amazonia. Neotrop. Primat. 6: 73-77. [ Links ]
Ford, S.M. 1986. Systematics of the New World monkeys. In D.R. Swindler & J. Erwing (eds.). Comparative primate biology. A.R. Liss, New York, Syst. Evol. Anat. 1: 73-135. [ Links ]
Ford, S.M. & L.C Davis. 1992. Systematics and body size. Implications for feeding adaptations in New World monkeys. Am. J. Phys. Anthropol. 88: 415-468. [ Links ]
Froehlich, J.W. & P.H. Froehlich. 1987. The status of Panamas endemic howling monkeys. Primat. Conserv. 8: 58-62. [ Links ]
Garcia, M.M., M.R. Cabballin, J. Aragones, C. Godai & J. Egozcue. 1975. Banding patterns of the chromosomes of Ateles geoffroyi with description of two cases of pericentric invertion. J. Med. Primatol. 4: 108-13. [ Links ]
Goloboff, P. 1993. PIWEE. Computer program and software. Tucuman, Argentina. (Available online: http://www.cladistics.com). [ Links ]
Gregorim, R. 1995. Variação geográfica e taxonomia das espécies brasileiras do gênero Alouatta Lacépède 1799 (Primates: Atelinae). MSc Dissertation Universidade de São Paulo São Paulo Brazil. [ Links ]
Groves, C.P. 1993. Primates. Cebidae. Alouattinae. In D.E. Wilson & D.R. Reeder. (eds.). Mammal species of the world. A taxonomic and geographic reference. Smithsonian Institute 254-255. [ Links ]
Hershkovitz, P. 1949. Mammals of Northern Colombia. Preliminary report No. 4. Monkeys Primates with taxonomic revisions of some forms. Proc U.S. Nat. Mus. 98: 323-470. [ Links ]
Hill, W.C.O. 1962. Primates. Comparative anatomy and taxonomy (Cebidae). Vol. V/Part B. Edinburgh University, Edinburgh. [ Links ]
Horovitz, I., R. Zardoya & A. Meyer. 1998. Platyrrhine systematics. a simultaneous analysis of molecular and morphological data. Am. J. Phys. Anthropol. 106: 261-281. [ Links ]
James, R.A., P.L. Leberg, J.M. Quattro & R.C. Vrijenhoek. 1997. Genetic diversity in black howler monkeys A. pigra from Belize. Am. J. Phys. Anthropol. 102: 329-336. [ Links ]
Kay, R.F. 1990. The phyletic relationships of extant and fossil Pitheciinae. J. Hum. Evol. 19: 175-208. [ Links ]
Kluge, A.G. 1989. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae: Serpentes). Syst. Zool. 38: 7-25. [ Links ]
Koiffman, C.P. 1982. Variabilidade cromossomica em macacos da familia Cebidae. In P.H. Saldanha. (Ed.). Genetica comparada de primatas Brasileiros. Sociedade Brasileira de Genetica Ribeirao Preto 13-132. [ Links ]
Koiffman, C.P. & P.H. Saldanha. 1974. Cytogenetics of Brazilian monkeys. J. Hum. Evol. 3: 275-282. [ Links ]
Kunkel, L.M., P.G. Heltne & D.S. Borgaonkar. 1980. Chromosomal variation and zoogeography in Ateles. Int. J. Primatol. 1: 223-232. [ Links ]
Lima, M.M.C. & H.N. Seuánez. 1991. Chromosome studies in the red howler monkey Alouatta seniculus stramineus (Platirrhini: Primates). description of an X1X2Y1Y2/X1X1X2X2 sex-chromosome system and karyological comparisons with other subspecies. Cytogen. Cell. Genet. 57: 151-156. [ Links ]
Lima, M.M.C., M.I.C. Sampaio, M.P.C. Schneider, W. Scheffrahn, H. Schneider & F.M. Salzano. 1990. Chromosomal and protein variation in the red howler monkeys. Rev. Brasil. Genet.13: 789-802. [ Links ]
Lima, M.M.C. & H.N. Seuánez. 1989. Cytogenetic characterization of Alouatta belzebul with atypical pelage coloration. Fol. Primatol. 52: 97-101. [ Links ]
Ma, N.S.F., T.C. Jones, R.W. Thorington, A. Miller & L. Morgan. 1975. Y-Autosome translocation in the Howler Monkey Alouatta palliata. J. Med. Primatol. 4: 299-307. [ Links ]
Madeiros, M.A., R.M.S. Barros, J.C. Pieczarka, C.Y. Nagamachi, M. Ponsa, M. Garcia, F. Garcia & J. Egozcue. 1997. Radiation and speciation of spider monkeys genus Ateles from a cytogenetic viewpoint. Int. J. Primatol. 42: 167-178. [ Links ]
Meireles, C.M., J. Czelusniak, S.F. Ferrari, M.P.C. Schneider & M. Goodman. 1999. Phylogenetic relationships among brazilian howler monkeys genus Alouatta (Platyrrhini: Atelidae) based on g1-globin pseudogene sequences. Genet Mol. Biol. 22: 337-344. [ Links ]
Minezawa, M., M. Harada, O.C. Jordan & C.J.V. Borda. 1985. Cytogenetics of Bolivian endemic red howler monkeys Alouatta seniculus sara. accsesory chromosomes and Y-autosome translocation related numerical variations. Kyoto Univ. Overs. Res. Rep. of New World Monk. 5: 7-16. [ Links ]
Mittermeier, R.A., A Rylands & A.F. Coimbra-Filho. 1988. Systematics. species ad subspecies- an update. In R.A. Mittermeier, A. Rylands, A.F. Coimbra-Filho & G.B. Fonseca (eds.). Ecology and behavior of neotropical primates vol. 2. World Wildlife Fundation Washington D.C. 13-75. [ Links ]
Mundy, N.I., A. Pissinatti & D.S. Woodruff. 2000. Multiple Nuclear Insertions of Mitochondrial Cytochrome b Sequences in Callitrichine (Primates). Mol. Biol. Evol. 17: 1075–1080. [ Links ]
Mudry, M., M. Ponsa, A Borrell, J. Egozcue & M. Garcia. 1994. Prometaphase chromosomes of the howler monkey Alouatta caraya. G, C, NOR and restriction enzyme REs banding. Am. J. Primatol. 33: 121-132. [ Links ]
Oliveira, E.H.C. 1996. Estudos citogeneticos e evolutivos nas especies brasileiras e argentinas do genero Alouatta Lacepede 1799 (Primates: Atelidae). MSc. Dissertation Universidade Federal do Parana Curitiba Brazil. [ Links ]
Oliveira, E.H.C., M.M.C. de Lima & I.J. Sbalqueiro. 1995. Chromosomal variation in Alouatta fusca. Neotrop. Primat. 3: 181-183. [ Links ]
Oliveira, E.H.C., M.M.C. de Lima, I.J. Sbalqueiro & A. Pissinatti. 1998. The karyotype of Alouatta fusca clamitans from Rio de Janeiro State Brazil. Evidence for Y-autosome translocation. Genet. Mol. Biol. 31: 361-364. [ Links ]
Péquinot, E.V., C.P. Koiffmann & B. Deutrillaux. 1985. Chormosomal phylogeny of Lagothrix Brachyteles and Cacajao. Cytogen. Cell. Genet. 39: 99-104. [ Links ]
Pieczarka, J.C., C.Y. Nagamashi & R.M.S. Barros. 1989. The kariotype of Ateles paniscus paniscus (Cebidae: Primates). 2n= 32. Rev. Brasil. Genet. 12: 543-551. [ Links ]
Pope, T.R. 1996. Socioecology population fragmentation and patterns of genetic loss. In J.C. Avise & J.L. Hamrick (ed.). Conservation genetics. Case histories from nature. Chapman and Hall. [ Links ]
Rosenberg, A.L. & K.B. Strier. 1989. Adaptive radiation of the Atelinae primates. J. Hum. Evol. 18: 171-750. [ Links ]
Roseberg, A.L. 1984. Fossil new world monkeys dispute the molecular clock. J. Hum. Evol. 13: 737-742. [ Links ]
Roseberg, A.L. 1981. Systematics. The higher taxa. A.F. Coimbra-Filho & R.A. Mittermeier (ed.). In Ecology and Behavior of neotropical primates. Rio de Janeiro. Academia Brasileira de Ciencias Vol. 1. 111-168. [ Links ]
Rylands, A.B., R.A. Mittermeier & E. Rodriquez-Luna. 1995. A species list for the New World primates Platyrrhini. Distribution by country endemism and conservation status according to the Mace-Land system. Neotrop. Primat. 3: 113-160. [ Links ]
Sampaio, I., M.P.C. Schneider & H. Schneider. 1996. Taxonomy of the Alouatta seniculus group. bio-chemical ad choromosome data. Primates 37: 65-73. [ Links ]
Schneider, H., M.P.C. Schneider, I. Sampaio, M.L. Harada, M. Stanhope, J. Czelusniak & M. Goodmoan. 1993. Molecular phylogeny of the New World monkeys Platyrrhini Primates. Mol. Phyl. Evol. 2: 225-242. [ Links ]
Schon-Ybarra, M.A. 1986. Loud calls of adult red howling monkeys Alouatta seniculus. Fol. Primatol. 47: 204-216. [ Links ]
Sites, J.W. & M. Craig. 1987. Chromosomal evolution and speciation revisited. Syst. Zool. 36: 153-174. [ Links ]
Stanyon, R., S. Tofanelli, M.A. Morescalchi, G. Agoramoorthy, O.A. Rayder & J. Wienberg. 1995. Cytogenetic analysis shows extensive genomic rearragements between Red Howler A. seniculus Linnaeus subspecies. Am. J. Primat. 35: 171-183. [ Links ]
Thompson, J.D., D.G. Higgins & T.J. Gibson. 1994. CLUSTAL W. improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nuc. Ac. Res. 22: 4673-4680. (Available online: http://www.ebi.ac.uk/clustalw/ ). [ Links ]
Vassart, M., A. Guedant, J.C. Vie, J. Keravec, A. Seguela & V.T. Volobouev. 1996. Chromosomes of Alouatta seniculus (Platyrrhini: Primates) from French Guiana. J. Heredit. 87: 331-334. [ Links ]
Viegas-Pequinot, E., C.P. Koiffmann & B. Dutrillaux. 1985. Chromosomal phylogeny of Lagothrix Brachyteles and Cacajao. Cytogenet. Cell. Genet. 39: 99-104. [ Links ]
White, M.J.D. 1978. Modes of Speciation. W. H. Freeman and Co. San Francisco USA pp. 45-106. [ Links ]
Yunis, E.J., O.M. Torres de Caballero, C. Ramirez & E. Ramirez. 1976. Chromosomal variations in the primate Alouatta seniculus seniculus. Fol. Prim. 25: 215-224. [ Links ]
Zingeser, M.R. 1973. Dentition of Brachyteles arachnoides with reference to to alouattine and atelinina affinities. Fol. Prim. 20: 351-390. [ Links ]
Karyotipic and morphological character description
Character 0. Number of metacentric chromosomes: with two metacentric pairs (0)-2, four and five metacentrics (1), six metacentrics (2), eight metacentrics (3), 10 metacentrics (4), 12 metacentrics (5).
Character 1. Number of submetacentric chromosomes: with six and eight submetacentrics (0), 10 submetacentrics (1), 12 submetacentrics (2), 14 submetacentrics (3), 16 submetacentrics (4), 18 submetacentrics (5), 20 submetacentrics (6), 22 submetacentrics (7).
Character 2. Number of acrocentric chromosomes: one or two acrocentrics (0), 15 acrocentrics (1), 22 or 24 acrocentrics (2), 26 or 27 acrocentrics (3), 28 acrocentrics (4), 30 or 32 acrocentrics (5), 36 acrocentrics (6).
Character 3. Diploid number: 0-32 y 34, 1- 43 y 44, 2-45 y 46, 3-47 y 48, 4-49, 5-50, 6-52, 7-53 y 54, 8-62.
Character 4. Presence of microchromosomes: absence (0), presence (1).
Character 5. Ordered. Type of duplication in male sex chromosome: simple (xy) (0), duplicated (xxyy) (1), partially duplicated (xxy) (2).
Chracater 6 Ordered. Type of female sex chromosomes: simple (XX) (0), duplicated (XXXX) (2).
Character 7. X chromosome morphololgy: acrocentric (0), metacentric (1), submetacentric (2).
Character 8. Ordered. Presence of Y chromosome translocations: absent (0), present (1).
Character 9. Ordered. Hioid volumen (ml) for male and female: 4.5/2 or less (0), 4.5/2 (1), 23-39/8-8.5 (2), 56.5-69.9/12.3- 12.5 (3).
Character 10. Male hioid bulla ventral shape: no developed (0), flat (1), round but not acute (2), round but acute (3), "w" shape (4).
Character 11. Male tentorium development: absent or no developed (0), rudimentario (1), evident without a chamber and slidely inflated (2), conspicous with an inflated chamber (3) .
Character 12. Male hioid shape aberture: without aperture (0), subrectangular (1), suboval (2), subpentameral with convex dorsal edge (3).
Character 13. Male hioid lateral shape: "l" shape (0), "j" shape (1), subtriangular (2), subrectangular (3).
Character 14. Ordered. Female hioid lateral shape: absent (0), round with curved edges in the last 1/3 (1), round with strait edges on the last 1/3 (2).
Character 15. Female tentorium dorsal part shape: absent (0), concavous (2), strait or flat (3).
Character 16. Female hioid aperture shape: without aperture (0), subrectangular (1), suboval (2), subpentameral (3).
Character 17. Ordered. Cornu development: reduced (0), prominent (1).
Character 18. Female hioid lateral shape: "l" shape (0), "j" shape (1), subtriangular (2), subrectangular (3).
Character 19. Ordered. Paraconule development: reduced (0), expanded (1).
Character 20. Ordered. Metaconule development: reduced (0), evident (1).
Character 21. Ordered. Presence of a folivorous diet: absent or infrequent (0), frecuent (1).
Character 22. Ordered. Occipital condile position: ventral (0), posterior (1).
Character 23. Ordered. Hioid shape: compresed plate like (0), chamber like (1).
Character 24. Ordered. Male hioid bulla development: absent (0), reduced (1), developed (2).
Character 25. Presence of male medial constriction on the hioid bone: absent (0), present (1).
Character 26. Male bulla apertures shape: developed and cuadrangular (0), oval (1), semicircular (2).
Character 27. Presence of a lateral furrow on the hioid bone: absent (0), present (1).
Character 28. Bulla dorsal wall inclination: expanded (0), strait (1).
Character 29. Ordered. Presence of the tentorium in males: absent (0), present (1).
Character 30. Male tentorium shape: concavous (0), convex and oval (1).
Character 31. Ordered. Male cornu development: developed (0), reduced (1).
Character 32. Ordered. Male hioid posterior shape: bowed (0), convex (1).
Character 33. Ordered. Male curniculum development: developed (0), reduced (1), absent (2).
Character 34. Ordered. Presence of female tentorium: absent (0), present (1).
Character 35. Female hiodeal bulla shape: rounded (0), elongated (1).
Character 36. Presence of a dorso-ventral constriction: absent (0), present (1).
Character 37. Female corniculum development: developed (0), reduced (1).
Character 38. Ordered. Presence of sexual dicromatism: absent (0), present (1).











 uBio
uBio